Note
Go to the end to download the full example code.
Aligned pipeline¶
In this example we will demonstrate use of AlignedUMAP with pipeline API.
Create Aligned Dataset¶
For using aligned UMAP, we need to use cartodata.pipeline.datasets.CSVSliceDataset. CSVSliceDataset divides the specified dataset into slice_count slices.
Setting the slice_type parameter to cumulative enables us to make slices cumulative.
Before slicing, we can sort the dataset using sort_asc_by parameter specifying the column name to sort in ascending order.
from cartodata.pipeline.datasets import CSVSliceDataset # noqa
from pathlib import Path # noqa
ROOT_DIR = Path.cwd().parent
# The directory where files necessary to load dataset columns reside
INPUT_DIR = ROOT_DIR / "datas"
# The directory where the generated dump files will be saved
TOP_DIR = ROOT_DIR / "dumps"
slice_count = 2
dataset = CSVSliceDataset(
"lisn", input_dir=INPUT_DIR, version="3.0.0", filename="lisn_2000_2022.csv",
fileurl="https://zenodo.org/record/7323538/files/lisn_2000_2022.csv",
columns=None, slice_count=slice_count, slice_type="discrete",
sort_asc="producedDateY_i", index_col=0
)
Now we should define our entities and set the column names corresponding to those entities from the data file. We have 4 entities:
———|-------------| | articles | en_title_s | | authors | authFullName_s | | labs | structAcronym_s | | words | en_abstract_s, en_title_s, en_keyword_s, en_domainAllCodeLabel_fs |
Cartolabe provides 4 types of columns:
IdentityColumn: The entity of this column represents the main entity of the dataset. The column data corresponding to the entity in the file should contain a single value and this value should be unique among column values. There can only be one IdentityColumn in the dataset.
CSColumn: The entity of this column type is related to the main entity, and can contain single or comma separated values.
CorpusColumn: The entity of this column type is the corpus related to the main entity. This can be a combination of multiple columns in the file. It uses a modified version of CountVectorizer(https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html#sklearn.feature_extraction.text.CountVectorizer).
TfidfCorpusColumn: The entity of this column type is the corpus related to the main entity. This can be a combination of multiple columns in the file or can contain filepath from which to read the text corpus. It uses TfidfVectorizer (https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.TfidfVectorizer.html).
In this dataset, Articles is our main entity. We will define it as IdentityColumn:
from cartodata.pipeline.columns import IdentityColumn, CSColumn, CorpusColumn # noqa
articles_column = IdentityColumn(nature="articles", column_name="en_title_s")
authors_column = CSColumn(
nature="authors", column_name="authFullName_s", filter_min_score=4
)
labs_column = CSColumn(
nature="labs", column_name="structAcronym_s", filter_min_score=4
)
words_column = CorpusColumn(
nature="words",
column_names=["en_abstract_s", "en_title_s",
"en_keyword_s", "en_domainAllCodeLabel_fs"],
stopwords="stopwords.txt", nb_grams=4, min_df=10, max_df=0.05,
min_word_length=5, normalize=True
)
dataset.set_columns(
[articles_column, authors_column, labs_column, words_column])
As we are going to use AlignedUMAP, we need to create an instance of cartodata.pipeline.common.AlignedPipeline.
from cartodata.pipeline.common import AlignedPipeline # noqa
pipeline = AlignedPipeline(dataset, top_dir=TOP_DIR, input_dir=INPUT_DIR)
Creating correspondance matrices for each entity type¶
We want to extract matrices that will map the correspondance between the articles and the entities we want to use.
AlignedPipeline has generate_entity_matrices function to generate matrices and scores for each entity (nature) specified for the dataset.
matrices_all, scores_all = pipeline.generate_entity_matrices()
We can list the sizes of each entity matrix for each slice.
for i in range(dataset.slice_count):
print(f"############## Slice {i + 1} #################")
matrices_i = matrices_all[i]
for nature, matrix in zip(pipeline.natures, matrices_i):
print(f"{nature} ------------- {matrix.shape}")
############## Slice 1 #################
articles ------------- (2131, 2131)
authors ------------- (2131, 328)
labs ------------- (2131, 314)
words ------------- (2131, 2506)
############## Slice 2 #################
articles ------------- (2131, 2131)
authors ------------- (2131, 396)
labs ------------- (2131, 387)
words ------------- (2131, 2850)
Dimension reduction¶
One way to see the matrices that we created is as coordinates in the space of all articles. What we want to do is to reduce the dimension of this space to make it easier to work with and see.
LSA projection
We’ll start by using the LSA (Latent Semantic Analysis) technique to reduce the number of rows in our data.
from cartodata.pipeline.projectionnd import LSAProjection # noqa
num_dim = 100
lsa_projection = LSAProjection(num_dim)
pipeline.set_projection_nd(lsa_projection)
Now we can run LSA projection on the matrices.
matrices_nD_all = pipeline.do_projection_nD()
""
for i in range(dataset.slice_count):
print(f"############## Slice {i + 1} #################")
matrices_nD = matrices_nD_all[i]
for nature, matrix in zip(pipeline.natures, matrices_nD):
print(f"{nature} ------------- {matrix.shape}")
############## Slice 1 #################
articles ------------- (100, 2131)
authors ------------- (100, 328)
labs ------------- (100, 314)
words ------------- (100, 2506)
############## Slice 2 #################
articles ------------- (100, 2131)
authors ------------- (100, 396)
labs ------------- (100, 387)
words ------------- (100, 2850)
This makes it easier to work with them for clustering or nearest neighbors tasks, but we also want to project them on a 2D space to be able to map them.
Aligned UMAP projection
The UMAP (Uniform Manifold Approximation and Projection) is a dimension reduction technique that can be used for visualisation similarly to t-SNE.
We use this algorithm to project matrices of each slice aligned with each other in 2 dimensions.
from cartodata.pipeline.projection2d import AlignedUMAPProjection # noqa
n_neighbors = 20
min_dists = 0.1
projection_2d = AlignedUMAPProjection(
n_neighbors=n_neighbors,
min_dist=min_dists,
init='random',
random_state=42,
n_epochs=200)
pipeline.set_projection_2d(projection_2d)
Now we can run AlignedUMAP projection on the LSA matrices.
matrices_2D_all, scores_final = pipeline.do_projection_2D()
""
for i in range(len(matrices_2D_all)):
print(f"############## Slice {i + 1} #################")
for j in range(len(matrices_2D_all[i])):
print(f"{matrices_2D_all[i][j].shape}")
/usr/local/lib/python3.9/site-packages/umap/umap_.py:1945: UserWarning: n_jobs value 1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/usr/local/lib/python3.9/site-packages/umap/umap_.py:1945: UserWarning: n_jobs value 1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1044: RuntimeWarning: overflow encountered in cast
epochs_per_sample[m].astype(np.float32) / negative_sample_rate
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1049: RuntimeWarning: overflow encountered in cast
epoch_of_next_sample.append(epochs_per_sample[m].astype(np.float32))
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1044: RuntimeWarning: overflow encountered in cast
epochs_per_sample[m].astype(np.float32) / negative_sample_rate
############## Slice 1 #################
(2, 2131)
(2, 328)
(2, 314)
(2, 2506)
############## Slice 2 #################
(2, 2131)
(2, 396)
(2, 387)
(2, 2850)
Now that we have 2D coordinates for our points, we can try to plot them to get a feel of the data’s shape.
labels = tuple(pipeline.natures)
colors = ['b', 'r', 'c', 'y', 'm']
for i in range(len(matrices_2D_all)):
matrices_2D = matrices_2D_all[i]
pipeline.plot_map(matrices_2D, labels, colors,
title=f"Slice {i+1}")
The plot above, as we don’t have labels for the points, doesn’t make much sense as is. But we can see that the data shows some clusters which we could try to identify.
Clustering¶
In order to identify clusters, we use the KMeans clustering technique on the articles. We’ll also try to label these clusters by selecting the most frequent words that appear in each cluster’s articles.
from cartodata.pipeline.clustering import KMeansClustering # noqa
# level of clusters, hl: high level, ml: medium level
cluster_natures = ["hl_clusters", "ml_clusters"]
kmeans_clustering = KMeansClustering(
n=8, base_factor=3, natures=cluster_natures)
pipeline.set_clustering(kmeans_clustering)
(clus_nD_all, clus_2D_all, clus_scores_all, clus_labels_all,
clus_eval_pos_all, clus_eval_neg_all) = pipeline.do_clustering()
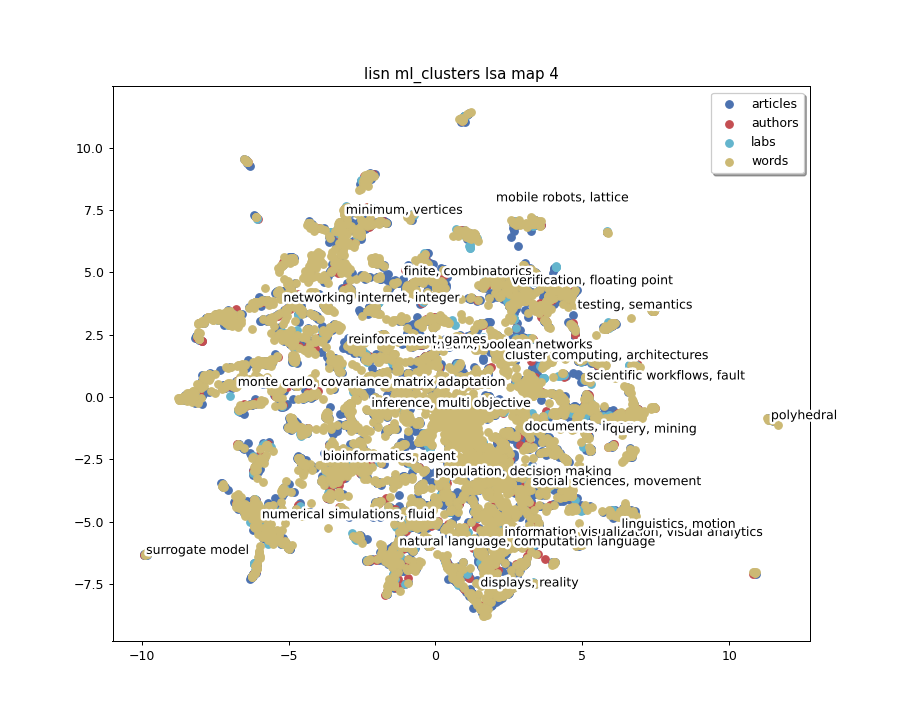
We will now display high level clusters:
for i in range(len(matrices_2D_all)):
ml_index = 1
clus_scores_ml = clus_scores_all[i][ml_index]
clus_mat_ml = clus_2D_all[i][ml_index]
fig_title = (
f"{pipeline.dataset.name} {pipeline.clustering.natures[ml_index]} "
f"{pipeline.projection_nd.key} slice {i + 1}/{pipeline.dataset.slice_count}"
)
matrices_2D = matrices_2D_all[i]
pipeline.plot_map(matrices_2D, labels, colors,
title=fig_title,
annotations=clus_scores_ml.index,
annotation_mat=clus_mat_ml)
Now we will save the plots in the working directory.
pipeline.save_plots()
[[(<Figure size 960x640 with 1 Axes>, 'lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_kmeans_hl_clusters_s1.png'), (<Figure size 960x640 with 1 Axes>, 'lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_kmeans_ml_clusters_s1.png')], [(<Figure size 960x640 with 1 Axes>, 'lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_kmeans_hl_clusters_s2.png'), (<Figure size 960x640 with 1 Axes>, 'lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_kmeans_ml_clusters_s2.png')]]
We set image name to view it later:
image_title_parts = pipeline.title_parts_clus("ml_clusters")
image_title_parts.append("s2")
image_name_ml_clusters_3_0_0 = "_".join(image_title_parts) + ".png"
image_name_ml_clusters_3_0_0
'lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_kmeans_ml_clusters_s2.png'
Nearest neighbors¶
One more thing which could be useful to appreciate the quality of our data would be to get each point’s nearest neighbors. If our data processing is done correctly, we expect the related articles, labs, words and authors to be located close to each other.
Finding nearest neighbors is a common task with various algorithms aiming to solve it. The find_neighboring method uses one of these algorithms to find the nearest points of all entities (articles, authors, labs, words). It takes an optional weight parameter to tweak the distance calculation to select points that have a higher score but are maybe a bit farther instead of just selecting the closest neighbors.
from cartodata.pipeline.neighbors import AllNeighbors # noqa
n_neighbors = 10
weights = [0, 0.5, 0.5, 0, 0]
neighboring = AllNeighbors(n_neighbors=n_neighbors, power_scores=weights)
pipeline.set_neighboring(neighboring)
pipeline.find_neighbors()
Export file using exporter¶
We can now export the data.
The exported data will be the points extracted from the dataset corresponding to the entities that we have defined.
In the export file, we will have the following columns for each point:
———|-------------| | nature | one of articles, authors, teams, labs, words | | label | point’s label | | score | point’s score | | rank | point’s rank | | x | point’s x location on the map | | y | point’s y location on the map | | nn_articles | neighboring articles to this point | | nn_labs | neighboring labs to this point | | nn_words | neighboring words to this point |
we will call pipeline.export function. It will create export.feather file and save under pipeline.working_dir.
from cartodata.pipeline.exporting import (
ExportNature, MetadataColumn
) # noqa
ex_author = ExportNature(key="authors",
refs=["labs"])
meta_year_article = MetadataColumn(column="producedDateY_i", as_column="year",
func="x.astype(str)")
meta_url_article = MetadataColumn(column="halId_s", as_column="url", func="x.fillna('')")
ex_article = ExportNature(key="articles", refs=["labs", "authors"],
add_metadata=[meta_year_article, meta_url_article])
""
pipeline.export(export_natures=[ex_article, ex_author])
Let’s see the directory structure for the pipeline’s working_dir:
for file in pipeline.working_dir.iterdir():
print(file)
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/authors_62027e49a8db161bscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/articles_62027e49a8db161bscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/authors_62027e49a8db161bscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/labs_62027e49a8db161bscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/labs_62027e49a8db161bscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/words_62027e49a8db161bscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/articles_62027e49a8db161bscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s1
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/words_62027e49a8db161bscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/lisn_2000_2022.csv
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/reducer_62027e49a8db161b.pkl
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/export.feather
Here we have directories s1 and s2 that contains the files for each slice. reducer.pkl file generated by serialization of the AlignedUMAP object. All scores files contain the scores for the last, ie. 2nd slice. These are saved to be used if a new version of dataset is to be aligned with this model.
Now we will view the contents of “s2”:
for file in (pipeline.working_dir / "s2").iterdir():
print(file)
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_s2.png
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/labs_aligned_umap.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/words_mat.npz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/words.index
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/articles_lsa.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_scores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_words_for_labs.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/authors_lsa.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_labs_for_authors.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_scores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/words.index.dat
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_articles_for_labs.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_lsa.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_authors_for_labs.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_eval_posscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_eval_negscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_authors_for_words.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_authors_for_authors.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/labs_lsa.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/labs_scores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_labels.json
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_aligned_umap.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_eval_posscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/authors_aligned_umap.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_eval_negscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/articles.index
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/authors_mat.npz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/words_scores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_eval_negscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/words_lsa.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_labs_for_words.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_aligned_umap.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_authors_for_articles.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_lsa.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/articles.index.dat
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/labs_scores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/authors_scores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/words_aligned_umap.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_kmeans_ml_clusters_s2.png
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/articles_scores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_articles_for_articles.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_labels.json
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_eval_posscores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/articles_aligned_umap.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/labs.index
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/lsa_components.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_articles_for_words.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_labs_for_articles.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_articles_for_authors.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_eval_negscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/authors.index
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/articles_mat.npz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/hl_clusters_scores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_eval_posscores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_words_for_words.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/words_scores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/lisn_3.0.0_45634fddf00a8a45_lsa_100_True_aligned_umap_euclidean_20_0.1_random_0.01_3_1.0_42_kmeans_hl_clusters_s2.png
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/labs_mat.npz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/ml_clusters_scores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/articles_scores.csv.bz2
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/authors_scores.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/authors.index.dat
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_words_for_articles.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_words_for_authors.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/labs.index.dat
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/nearest_labs_for_labs.npy.gz
/builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.0.0/s2/export.feather
Let’s display the contents of the export.feather file under d2 directory for the 2nd slice of the dataset.
import pandas as pd # noqa
df = pd.read_feather(pipeline.working_dir / "s2" / "export.feather")
df.head()
We will store the matrices ad clusters to be used later for an animation to see the evolution of the map.
clus_scores_plt = clus_scores_all
clus_2D_plt = clus_2D_all
matrices_2D_plt = matrices_2D_all
Align new version of dataset¶
We have sliced version 3.0.0 of LISN dataset into 2 and created 2D projections aligned with each other. It contains data until year 2022 inclusive.
Now we will download recent version of LISN data that contains publications of year 2023 as well and we will align it with version 3.0.0 of LISN dataset.
from cartodata.scraping import scrape_hal, process_domain_column # noqa
from cartodata.command_line import STRUCT_MAP # noqa
filters = {}
struct = "lisn"
yearfrom = 2000
yearto = 2024
file = dataset.input_dir / f"{struct.lower()}_{yearfrom}_{yearto - 1}.csv"
if not file.exists():
filters['structId_i'] = "(" + STRUCT_MAP[struct] + ")"
years = range(yearfrom, yearto)
df = scrape_hal(struct, filters, years, cool_down=2)
process_domain_column(df)
df.to_csv(file)
2024-10-03 14:21:58,782 - cartodata.scraping - DEBUG - Starting mining task from HAL...
2024-10-03 14:22:00,984 - cartodata.scraping - DEBUG - Importing 22 docs for year 2000
/builds/2mk6rsew/0/hgozukan/cartolabe-data/cartodata/scraping.py:116: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation.
df = pd.concat([df, year_df])
2024-10-03 14:22:04,373 - cartodata.scraping - DEBUG - Importing 17 docs for year 2001
2024-10-03 14:22:08,724 - cartodata.scraping - DEBUG - Importing 26 docs for year 2002
2024-10-03 14:22:13,092 - cartodata.scraping - DEBUG - Importing 44 docs for year 2003
2024-10-03 14:22:16,477 - cartodata.scraping - DEBUG - Importing 90 docs for year 2004
2024-10-03 14:22:18,880 - cartodata.scraping - DEBUG - Importing 95 docs for year 2005
2024-10-03 14:22:22,323 - cartodata.scraping - DEBUG - Importing 148 docs for year 2006
2024-10-03 14:22:24,811 - cartodata.scraping - DEBUG - Importing 151 docs for year 2007
2024-10-03 14:22:28,264 - cartodata.scraping - DEBUG - Importing 199 docs for year 2008
2024-10-03 14:22:31,721 - cartodata.scraping - DEBUG - Importing 284 docs for year 2009
2024-10-03 14:22:36,237 - cartodata.scraping - DEBUG - Importing 335 docs for year 2010
2024-10-03 14:22:39,825 - cartodata.scraping - DEBUG - Importing 350 docs for year 2011
2024-10-03 14:22:43,374 - cartodata.scraping - DEBUG - Importing 352 docs for year 2012
2024-10-03 14:22:46,884 - cartodata.scraping - DEBUG - Importing 402 docs for year 2013
2024-10-03 14:22:51,431 - cartodata.scraping - DEBUG - Importing 402 docs for year 2014
2024-10-03 14:22:54,951 - cartodata.scraping - DEBUG - Importing 406 docs for year 2015
2024-10-03 14:22:57,432 - cartodata.scraping - DEBUG - Importing 439 docs for year 2016
2024-10-03 14:23:00,974 - cartodata.scraping - DEBUG - Importing 396 docs for year 2017
2024-10-03 14:23:04,537 - cartodata.scraping - DEBUG - Importing 417 docs for year 2018
2024-10-03 14:23:08,090 - cartodata.scraping - DEBUG - Importing 396 docs for year 2019
2024-10-03 14:23:11,672 - cartodata.scraping - DEBUG - Importing 320 docs for year 2020
2024-10-03 14:23:15,260 - cartodata.scraping - DEBUG - Importing 481 docs for year 2021
2024-10-03 14:23:18,865 - cartodata.scraping - DEBUG - Importing 543 docs for year 2022
2024-10-03 14:23:22,441 - cartodata.scraping - DEBUG - Importing 569 docs for year 2023
At this point we have two options to align with version 3.0.0.
We can use current projection_2d instance and update using that.
We can initiate a new AlignedUMAP instance and align specifying the previous version.
Using current AlignedUMAP
We will create a new dataset for the new dataset file. We will version this as 3.1.0.
slice_count = 1
new_dataset = CSVSliceDataset(
"lisn", input_dir=INPUT_DIR, filename="lisn_2000_2023.csv",
slice_count=slice_count, version="3.1.0", sort_asc="producedDateY_i"
)
new_dataset.set_columns(
[articles_column, authors_column, labs_column, words_column])
We will update pipeline’s dataset with the new one.
pipeline.update_dataset(new_dataset)
We will generate entity matrices for the new dataset.
matrices_all, scores_all = pipeline.generate_entity_matrices()
2024-10-03 14:23:24,638 - cartodata.pipeline.datasets - INFO - Generating entity matrices...
2024-10-03 14:23:24,639 - cartodata.pipeline.base - INFO - Entity matrix for nature articles does not exist for mat.
2024-10-03 14:23:24,639 - cartodata.pipeline.columns - INFO - Loading articles nature...
TIMEIT 'load_identity_column' 0.81 ms
2024-10-03 14:23:24,640 - cartodata.pipeline.columns - INFO - Loading authors nature...
TIMEIT 'load_comma_separated_column' 239.49 ms
2024-10-03 14:23:24,880 - cartodata.pipeline.columns - INFO - The matrix has 8312 columns. Filtering authors with less than 4 entities.
2024-10-03 14:23:24,885 - cartodata.pipeline.columns - INFO - Working with 783 authors.
2024-10-03 14:23:24,885 - cartodata.pipeline.columns - INFO - Loading labs nature...
TIMEIT 'load_comma_separated_column' 330.58 ms
2024-10-03 14:23:25,216 - cartodata.pipeline.columns - INFO - The matrix has 2138 columns. Filtering labs with less than 4 entities.
2024-10-03 14:23:25,220 - cartodata.pipeline.columns - INFO - Working with 677 labs.
2024-10-03 14:23:25,220 - cartodata.pipeline.columns - INFO - Loading words nature...
TIMEIT 'load_text_column' 12.34 s
2024-10-03 14:23:37,801 - cartodata.pipeline.datasets - INFO - Entity matrices generated.
2024-10-03 14:23:37,802 - cartodata.operations - INFO - Saving articles score vector of length 4865 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/articles_scores.
2024-10-03 14:23:37,871 - cartodata.operations - INFO - Saving authors score vector of length 783 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/authors_scores.
2024-10-03 14:23:37,881 - cartodata.operations - INFO - Saving labs score vector of length 677 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/labs_scores.
2024-10-03 14:23:37,888 - cartodata.operations - INFO - Saving words score vector of length 5257 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/words_scores.
2024-10-03 14:23:37,951 - cartodata.pipeline.base - INFO - Saving matrices for mat ...
2024-10-03 14:23:37,954 - cartodata.operations - INFO - Saving articles matrix of size (4865, 4865) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/articles_mat.npz.
2024-10-03 14:23:37,962 - cartodata.operations - INFO - Saving authors matrix of size (4865, 783) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/authors_mat.npz.
2024-10-03 14:23:37,977 - cartodata.operations - INFO - Saving labs matrix of size (4865, 677) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/labs_mat.npz.
2024-10-03 14:23:38,163 - cartodata.operations - INFO - Saving words matrix of size (4865, 5257) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/words_mat.npz.
We will do LSA projection.
from cartodata.pipeline.projectionnd import LSAProjection # noqa
num_dim = 100
lsa_projection = LSAProjection(num_dim)
pipeline.set_projection_nd(lsa_projection)
matrices_nD_all = pipeline.do_projection_nD()
""
matrices_nD = matrices_nD_all[0]
for nature, matrix in zip(pipeline.natures, matrices_nD):
print(f"{nature} ------------- {matrix.shape}")
2024-10-03 14:23:38,167 - cartodata.pipeline.base - INFO - Entity matrix for nature articles does not exist for lsa.
2024-10-03 14:23:38,167 - cartodata.pipeline.base - INFO - Loading mat matrices from dumps...
2024-10-03 14:23:38,171 - cartodata.operations - INFO - Loaded articles matrix of size (4865, 4865).
2024-10-03 14:23:38,176 - cartodata.operations - INFO - Loaded authors matrix of size (4865, 783).
2024-10-03 14:23:38,184 - cartodata.operations - INFO - Loaded labs matrix of size (4865, 677).
2024-10-03 14:23:38,210 - cartodata.operations - INFO - Loaded words matrix of size (4865, 5257).
2024-10-03 14:23:38,210 - cartodata.pipeline.projectionnd - INFO - Starting lsa projection...
TIMEIT 'lsa_projection' 1.11 s
2024-10-03 14:23:39,318 - cartodata.pipeline.base - INFO - Saving matrices for lsa ...
2024-10-03 14:23:39,554 - cartodata.operations - INFO - Saving articles matrix of size (100, 4865) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/articles_lsa.npy.gz.
2024-10-03 14:23:39,591 - cartodata.operations - INFO - Saving authors matrix of size (100, 783) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/authors_lsa.npy.gz.
2024-10-03 14:23:39,623 - cartodata.operations - INFO - Saving labs matrix of size (100, 677) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/labs_lsa.npy.gz.
2024-10-03 14:23:39,871 - cartodata.operations - INFO - Saving words matrix of size (100, 5257) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/words_lsa.npy.gz.
2024-10-03 14:23:39,871 - cartodata.pipeline.projectionnd - INFO - Finished lsa projection
articles ------------- (100, 4865)
authors ------------- (100, 783)
labs ------------- (100, 677)
words ------------- (100, 5257)
Now we will run aligned UMAP to align the current version 3.1.0 of dataset with the current reducer.
matrices_2D_all, scores_final = pipeline.do_projection_2D(prev_version="3.0.0")
""
for i in range(len(matrices_2D_all[0])):
print(f"{matrices_2D_all[0][j].shape}")
""
labels = tuple(pipeline.natures)
matrices_2D = matrices_2D_all[0]
pipeline.plot_map(matrices_2D, labels, colors,
title="New Dataset")
""
(clus_nD_all, clus_2D_all, clus_scores_all, clus_labels_all,
clus_eval_pos_all, clus_eval_neg_all) = pipeline.do_clustering()
""
ml_index = 1
clus_scores_ml = clus_scores_all[0][ml_index]
clus_mat_ml = clus_2D_all[0][ml_index]
fig_title = (
f"{pipeline.dataset.name} {pipeline.clustering.natures[ml_index]} "
f"{pipeline.projection_nd.key} New Dataset"
)
matrices_2D = matrices_2D_all[0]
pipeline.plot_map(matrices_2D, labels, colors,
title=fig_title,
annotations=clus_scores_ml.index,
annotation_mat=clus_mat_ml)
""
pipeline.save_plots()
""
clus_scores_plt += clus_scores_all
clus_2D_plt += clus_2D_all
matrices_2D_plt += matrices_2D_all
2024-10-03 14:23:39,875 - cartodata.pipeline.base - INFO - Entity matrix for nature articles does not exist for aligned_umap.
2024-10-03 14:23:39,875 - cartodata.pipeline.base - INFO - Loading lsa matrices from dumps...
2024-10-03 14:23:39,912 - cartodata.operations - INFO - Loaded articles matrix of size (100, 4865).
2024-10-03 14:23:39,919 - cartodata.operations - INFO - Loaded authors matrix of size (100, 783).
2024-10-03 14:23:39,926 - cartodata.operations - INFO - Loaded labs matrix of size (100, 677).
2024-10-03 14:23:39,965 - cartodata.operations - INFO - Loaded words matrix of size (100, 5257).
2024-10-03 14:23:40,046 - cartodata.operations - INFO - Loaded articles score vector of length 4865.
2024-10-03 14:23:40,052 - cartodata.operations - INFO - Loaded authors score vector of length 783.
2024-10-03 14:23:40,057 - cartodata.operations - INFO - Loaded labs score vector of length 677.
2024-10-03 14:23:40,071 - cartodata.operations - INFO - Loaded words score vector of length 5257.
2024-10-03 14:23:40,107 - cartodata.operations - INFO - Loaded articles score vector of length 2131.
2024-10-03 14:23:40,112 - cartodata.operations - INFO - Loaded authors score vector of length 396.
2024-10-03 14:23:40,116 - cartodata.operations - INFO - Loaded labs score vector of length 387.
2024-10-03 14:23:40,124 - cartodata.operations - INFO - Loaded words score vector of length 2850.
/usr/local/lib/python3.9/site-packages/umap/umap_.py:1945: UserWarning: n_jobs value 1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/usr/local/lib/python3.9/site-packages/umap/aligned_umap.py:195: NumbaTypeSafetyWarning: unsafe cast from int64 to int32. Precision may be lost.
if i in relation_dict:
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1044: RuntimeWarning: overflow encountered in cast
epochs_per_sample[m].astype(np.float32) / negative_sample_rate
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1049: RuntimeWarning: overflow encountered in cast
epoch_of_next_sample.append(epochs_per_sample[m].astype(np.float32))
2024-10-03 14:26:18,894 - cartodata.pipeline.base - INFO - Saving matrices for aligned_umap ...
2024-10-03 14:26:18,896 - cartodata.operations - INFO - Saving articles matrix of size (2, 4865) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/articles_aligned_umap.npy.gz.
2024-10-03 14:26:18,897 - cartodata.operations - INFO - Saving authors matrix of size (2, 783) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/authors_aligned_umap.npy.gz.
2024-10-03 14:26:18,898 - cartodata.operations - INFO - Saving labs matrix of size (2, 677) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/labs_aligned_umap.npy.gz.
2024-10-03 14:26:18,901 - cartodata.operations - INFO - Saving words matrix of size (2, 5257) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/words_aligned_umap.npy.gz.
2024-10-03 14:26:18,901 - cartodata.operations - INFO - Saving articles score vector of length 4865 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/articles_62027e49a8db161bscores.
2024-10-03 14:26:18,968 - cartodata.operations - INFO - Saving authors score vector of length 783 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/authors_62027e49a8db161bscores.
2024-10-03 14:26:18,976 - cartodata.operations - INFO - Saving labs score vector of length 677 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/labs_62027e49a8db161bscores.
2024-10-03 14:26:18,983 - cartodata.operations - INFO - Saving words score vector of length 5257 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/words_62027e49a8db161bscores.
(2, 5257)
(2, 5257)
(2, 5257)
(2, 5257)
2024-10-03 14:26:19,089 - cartodata.pipeline.base - INFO - Entity matrix for nature hl_clusters does not exist for kmeans.
2024-10-03 14:26:19,090 - cartodata.pipeline.base - INFO - Loading mat matrices from dumps...
2024-10-03 14:26:19,093 - cartodata.operations - INFO - Loaded articles matrix of size (4865, 4865).
2024-10-03 14:26:19,099 - cartodata.operations - INFO - Loaded authors matrix of size (4865, 783).
2024-10-03 14:26:19,106 - cartodata.operations - INFO - Loaded labs matrix of size (4865, 677).
2024-10-03 14:26:19,131 - cartodata.operations - INFO - Loaded words matrix of size (4865, 5257).
2024-10-03 14:26:19,207 - cartodata.operations - INFO - Loaded articles score vector of length 4865.
2024-10-03 14:26:19,213 - cartodata.operations - INFO - Loaded authors score vector of length 783.
2024-10-03 14:26:19,217 - cartodata.operations - INFO - Loaded labs score vector of length 677.
2024-10-03 14:26:19,229 - cartodata.operations - INFO - Loaded words score vector of length 5257.
2024-10-03 14:26:19,230 - cartodata.pipeline.base - INFO - Loading lsa matrices from dumps...
2024-10-03 14:26:19,266 - cartodata.operations - INFO - Loaded articles matrix of size (100, 4865).
2024-10-03 14:26:19,273 - cartodata.operations - INFO - Loaded authors matrix of size (100, 783).
2024-10-03 14:26:19,279 - cartodata.operations - INFO - Loaded labs matrix of size (100, 677).
2024-10-03 14:26:19,318 - cartodata.operations - INFO - Loaded words matrix of size (100, 5257).
2024-10-03 14:26:19,318 - cartodata.pipeline.base - INFO - Loading aligned_umap matrices from dumps...
2024-10-03 14:26:19,320 - cartodata.operations - INFO - Loaded articles matrix of size (2, 4865).
2024-10-03 14:26:19,321 - cartodata.operations - INFO - Loaded authors matrix of size (2, 783).
2024-10-03 14:26:19,322 - cartodata.operations - INFO - Loaded labs matrix of size (2, 677).
2024-10-03 14:26:19,324 - cartodata.operations - INFO - Loaded words matrix of size (2, 5257).
2024-10-03 14:26:19,324 - cartodata.pipeline.clustering - INFO - Starting kmeans clustering for hl_clusters. Nb clusters = 8.
TIMEIT 'create_kmeans_clusters' 107.15 ms
2024-10-03 14:26:19,433 - cartodata.pipeline.clustering - INFO - Starting kmeans clustering for ml_clusters. Nb clusters = 24.
TIMEIT 'create_kmeans_clusters' 250.59 ms
2024-10-03 14:26:19,685 - cartodata.operations - INFO - Saving hl_clusters score vector of length 8 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/hl_clusters_scores.
2024-10-03 14:26:19,687 - cartodata.operations - INFO - Saving ml_clusters score vector of length 24 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/ml_clusters_scores.
2024-10-03 14:26:19,690 - cartodata.pipeline.base - INFO - Saving matrices for lsa ...
2024-10-03 14:26:19,691 - cartodata.operations - INFO - Saving hl_clusters matrix of size (100, 8) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/hl_clusters_lsa.npy.gz.
2024-10-03 14:26:19,692 - cartodata.operations - INFO - Saving ml_clusters matrix of size (100, 24) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/ml_clusters_lsa.npy.gz.
2024-10-03 14:26:19,692 - cartodata.pipeline.base - INFO - Saving matrices for aligned_umap ...
2024-10-03 14:26:19,693 - cartodata.operations - INFO - Saving hl_clusters matrix of size (2, 8) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/hl_clusters_aligned_umap.npy.gz.
2024-10-03 14:26:19,693 - cartodata.operations - INFO - Saving ml_clusters matrix of size (2, 24) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/ml_clusters_aligned_umap.npy.gz.
2024-10-03 14:26:19,694 - cartodata.operations - INFO - Saving hl_clusters cluster labels into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/hl_clusters_labels.json
2024-10-03 14:26:19,726 - cartodata.operations - INFO - Saving ml_clusters cluster labels into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/ml_clusters_labels.json
2024-10-03 14:26:19,793 - cartodata.operations - INFO - Saving hl_clusters score vector of length 8 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/hl_clusters_eval_posscores.
2024-10-03 14:26:19,796 - cartodata.operations - INFO - Saving ml_clusters score vector of length 24 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/ml_clusters_eval_posscores.
2024-10-03 14:26:19,799 - cartodata.operations - INFO - Saving hl_clusters score vector of length 8 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/hl_clusters_eval_negscores.
2024-10-03 14:26:19,801 - cartodata.operations - INFO - Saving ml_clusters score vector of length 24 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/ml_clusters_eval_negscores.
2024-10-03 14:26:19,856 - cartodata.pipeline.base - INFO - Loading aligned_umap matrices from dumps...
2024-10-03 14:26:19,858 - cartodata.operations - INFO - Loaded articles matrix of size (2, 4865).
2024-10-03 14:26:19,859 - cartodata.operations - INFO - Loaded authors matrix of size (2, 783).
2024-10-03 14:26:19,860 - cartodata.operations - INFO - Loaded labs matrix of size (2, 677).
2024-10-03 14:26:19,862 - cartodata.operations - INFO - Loaded words matrix of size (2, 5257).
2024-10-03 14:26:20,200 - cartodata.operations - INFO - Loaded hl_clusters score vector of length 8.
2024-10-03 14:26:20,204 - cartodata.operations - INFO - Loaded ml_clusters score vector of length 24.
2024-10-03 14:26:20,204 - cartodata.pipeline.base - INFO - Loading lsa matrices from dumps...
2024-10-03 14:26:20,205 - cartodata.operations - INFO - Loaded hl_clusters matrix of size (100, 8).
2024-10-03 14:26:20,206 - cartodata.operations - INFO - Loaded ml_clusters matrix of size (100, 24).
2024-10-03 14:26:20,207 - cartodata.pipeline.base - INFO - Loading aligned_umap matrices from dumps...
2024-10-03 14:26:20,208 - cartodata.operations - INFO - Loaded hl_clusters matrix of size (2, 8).
2024-10-03 14:26:20,209 - cartodata.operations - INFO - Loaded ml_clusters matrix of size (2, 24).
2024-10-03 14:26:20,214 - cartodata.operations - INFO - Loaded hl_clusters score vector of length 8.
2024-10-03 14:26:20,217 - cartodata.operations - INFO - Loaded ml_clusters score vector of length 24.
2024-10-03 14:26:20,222 - cartodata.operations - INFO - Loaded hl_clusters score vector of length 8.
2024-10-03 14:26:20,225 - cartodata.operations - INFO - Loaded ml_clusters score vector of length 24.
2024-10-03 14:26:20,225 - cartodata.operations - INFO - Loaded hl_clusters cluster labels from file /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/hl_clusters_labels.json.
2024-10-03 14:26:20,232 - cartodata.operations - INFO - Loaded ml_clusters cluster labels from file /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.1.0/s1/ml_clusters_labels.json.
Let’s view the dataset version 3.1.0 with the final slice of the dataset version 3.0.0.
import matplotlib.pyplot as plt # noqa
image_title_parts = pipeline.title_parts_clus("ml_clusters")
image_title_parts.append("s1")
image_name_ml_clusters_3_1_0 = "_".join(image_title_parts) + ".png"
img1 = plt.imread(pipeline.working_dir.parent / "3.0.0" / "s2" / image_name_ml_clusters_3_0_0)
img2 = plt.imread(pipeline.working_dir / "s1" / image_name_ml_clusters_3_1_0)
f, ax = plt.subplots(2, 1, figsize=(9, 12))
ax[0].imshow(img1)
ax[1].imshow(img2)
ax[0].axis('off')
ax[1].axis('off')
plt.tight_layout()
plt.show()

Using new AlignedUmap instance
Now we will follow the alternative flow and create a new instance of AlignedUMAPProjection and load the reducer from version 3.0.0 by reading the reducer.pkl file.
First we will create new dataset, with version 3.2.0 with the same data.
new_dataset = CSVSliceDataset(
"lisn", input_dir=INPUT_DIR, filename="lisn_2000_2023.csv",
slice_count=slice_count, version="3.2.0", sort_asc="producedDateY_i"
)
new_dataset.set_columns(
[articles_column, authors_column, labs_column, words_column])
pipeline.update_dataset(new_dataset)
""
n_neighbors = 20
min_dists = 0.1
projection_2d = AlignedUMAPProjection(
n_neighbors=n_neighbors,
min_dist=min_dists,
init='random',
random_state=42,
n_epochs=200)
pipeline.set_projection_2d(projection_2d)
""
matrices_all, scores_all = pipeline.generate_entity_matrices()
""
from cartodata.pipeline.projectionnd import LSAProjection # noqa
num_dim = 100
lsa_projection = LSAProjection(num_dim)
pipeline.set_projection_nd(lsa_projection)
matrices_nD_all = pipeline.do_projection_nD()
""
matrices_2D_all, scores_final = pipeline.do_projection_2D(prev_version="3.0.0")
""
for i in range(len(matrices_2D_all[0])):
print(f"{matrices_2D_all[0][j].shape}")
""
labels = tuple(pipeline.natures)
matrices_2D = matrices_2D_all[0]
pipeline.plot_map(matrices_2D, labels, colors, title="New Dataset")
""
(clus_nD_all, clus_2D_all, clus_scores_all, clus_labels_all,
clus_eval_pos_all, clus_eval_neg_all) = pipeline.do_clustering()
""
ml_index = 1
clus_scores_ml = clus_scores_all[0][ml_index]
clus_mat_ml = clus_2D_all[0][ml_index]
fig_title = (
f"{pipeline.dataset.name} {pipeline.clustering.natures[ml_index]} "
f"{pipeline.projection_nd.key} New Dataset"
)
matrices_2D = matrices_2D_all[0]
pipeline.plot_map(matrices_2D, labels, colors, title=fig_title,
annotations=clus_scores_ml.index,
annotation_mat=clus_mat_ml)
""
pipeline.save_plots()
""
import matplotlib.pyplot as plt # noqa
image_title_parts = pipeline.title_parts_clus("ml_clusters")
image_title_parts.append("s1")
image_name_ml_clusters_3_2_0 = "_".join(image_title_parts) + ".png"
img1 = plt.imread(pipeline.working_dir.parent / "3.0.0" / "s2" / image_name_ml_clusters_3_0_0)
img2 = plt.imread(pipeline.working_dir.parent/ "3.1.0" / "s1" / image_name_ml_clusters_3_1_0)
img3 = plt.imread(pipeline.working_dir / "s1" / image_name_ml_clusters_3_2_0)
f, ax = plt.subplots(3, 1, figsize=(20, 15))
ax[0].imshow(img1)
ax[1].imshow(img2)
ax[2].imshow(img3)
ax[0].axis('off')
ax[1].axis('off')
ax[2].axis('off')
plt.tight_layout()
plt.show()
""
clus_scores_plt += clus_scores_all
clus_2D_plt += clus_2D_all
matrices_2D_plt += matrices_2D_all
""
from matplotlib import animation
import matplotlib.patheffects as pe # noqa
fig = plt.figure(figsize=[10,8], frameon=False, dpi=90)
ax = plt.gca()
def plot_map(matrices, labels, title=None,
annotations=None, annotation_mat=None, annotation_color='black'):
ax.clear()
axes = []
for i, m in enumerate(matrices):
axes.append(ax.scatter(m[0, :], m[1, :],
color=colors[i],
label=labels[i]))
# set title
if title is not None:
ax.set_title(title)
# set legend
ax.legend(tuple(axes), labels, fancybox=True, shadow=True)
if annotations is not None and annotation_mat is not None:
for i in range(len(annotations)):
ax.annotate(annotations[i],
(annotation_mat[0, i], annotation_mat[1, i]),
color=annotation_color,
path_effects=[
pe.withStroke(linewidth=4, foreground="white")
])
return axes
def animate(i):
ml_index = 1
clus_scores_ml = clus_scores_plt[i][ml_index]
clus_mat_ml = clus_2D_plt[i][ml_index]
fig_title = (
f"{pipeline.dataset.name} {pipeline.clustering.natures[ml_index]} "
f"{pipeline.projection_nd.key} map {i + 1}"
)
matrices_2D = matrices_2D_plt[i]
return plot_map(matrices_2D, labels, title=fig_title,
annotations=clus_scores_ml.index,
annotation_mat=clus_mat_ml)
anim = animation.FuncAnimation(fig, animate, repeat = True,
frames=range(0,len(matrices_2D_plt)), interval=3000,
blit=True, repeat_delay=3000)
plt.show()
""
anim.save("anim.gif", writer="imagemagick",fps=1, bitrate=100, dpi=80)
""
from IPython.display import Image
Image(filename='anim.gif')
2024-10-03 14:26:23,934 - cartodata.pipeline.datasets - INFO - Generating entity matrices...
2024-10-03 14:26:23,934 - cartodata.pipeline.base - INFO - Entity matrix for nature articles does not exist for mat.
2024-10-03 14:26:23,935 - cartodata.pipeline.columns - INFO - Loading articles nature...
TIMEIT 'load_identity_column' 0.72 ms
2024-10-03 14:26:23,936 - cartodata.pipeline.columns - INFO - Loading authors nature...
TIMEIT 'load_comma_separated_column' 231.05 ms
2024-10-03 14:26:24,167 - cartodata.pipeline.columns - INFO - The matrix has 8312 columns. Filtering authors with less than 4 entities.
2024-10-03 14:26:24,172 - cartodata.pipeline.columns - INFO - Working with 783 authors.
2024-10-03 14:26:24,172 - cartodata.pipeline.columns - INFO - Loading labs nature...
TIMEIT 'load_comma_separated_column' 331.71 ms
2024-10-03 14:26:24,505 - cartodata.pipeline.columns - INFO - The matrix has 2138 columns. Filtering labs with less than 4 entities.
2024-10-03 14:26:24,508 - cartodata.pipeline.columns - INFO - Working with 677 labs.
2024-10-03 14:26:24,508 - cartodata.pipeline.columns - INFO - Loading words nature...
TIMEIT 'load_text_column' 12.50 s
2024-10-03 14:26:37,247 - cartodata.pipeline.datasets - INFO - Entity matrices generated.
2024-10-03 14:26:37,248 - cartodata.operations - INFO - Saving articles score vector of length 4865 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/articles_scores.
2024-10-03 14:26:37,317 - cartodata.operations - INFO - Saving authors score vector of length 783 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/authors_scores.
2024-10-03 14:26:37,326 - cartodata.operations - INFO - Saving labs score vector of length 677 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/labs_scores.
2024-10-03 14:26:37,333 - cartodata.operations - INFO - Saving words score vector of length 5257 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/words_scores.
2024-10-03 14:26:37,396 - cartodata.pipeline.base - INFO - Saving matrices for mat ...
2024-10-03 14:26:37,398 - cartodata.operations - INFO - Saving articles matrix of size (4865, 4865) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/articles_mat.npz.
2024-10-03 14:26:37,405 - cartodata.operations - INFO - Saving authors matrix of size (4865, 783) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/authors_mat.npz.
2024-10-03 14:26:37,420 - cartodata.operations - INFO - Saving labs matrix of size (4865, 677) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/labs_mat.npz.
2024-10-03 14:26:37,605 - cartodata.operations - INFO - Saving words matrix of size (4865, 5257) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/words_mat.npz.
2024-10-03 14:26:37,610 - cartodata.pipeline.base - INFO - Entity matrix for nature articles does not exist for lsa.
2024-10-03 14:26:37,610 - cartodata.pipeline.base - INFO - Loading mat matrices from dumps...
2024-10-03 14:26:37,614 - cartodata.operations - INFO - Loaded articles matrix of size (4865, 4865).
2024-10-03 14:26:37,619 - cartodata.operations - INFO - Loaded authors matrix of size (4865, 783).
2024-10-03 14:26:37,626 - cartodata.operations - INFO - Loaded labs matrix of size (4865, 677).
2024-10-03 14:26:37,651 - cartodata.operations - INFO - Loaded words matrix of size (4865, 5257).
2024-10-03 14:26:37,651 - cartodata.pipeline.projectionnd - INFO - Starting lsa projection...
TIMEIT 'lsa_projection' 1.08 s
2024-10-03 14:26:38,732 - cartodata.pipeline.base - INFO - Saving matrices for lsa ...
2024-10-03 14:26:38,967 - cartodata.operations - INFO - Saving articles matrix of size (100, 4865) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/articles_lsa.npy.gz.
2024-10-03 14:26:39,004 - cartodata.operations - INFO - Saving authors matrix of size (100, 783) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/authors_lsa.npy.gz.
2024-10-03 14:26:39,036 - cartodata.operations - INFO - Saving labs matrix of size (100, 677) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/labs_lsa.npy.gz.
2024-10-03 14:26:39,285 - cartodata.operations - INFO - Saving words matrix of size (100, 5257) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/words_lsa.npy.gz.
2024-10-03 14:26:39,286 - cartodata.pipeline.projectionnd - INFO - Finished lsa projection
2024-10-03 14:26:39,287 - cartodata.pipeline.base - INFO - Entity matrix for nature articles does not exist for aligned_umap.
2024-10-03 14:26:39,287 - cartodata.pipeline.base - INFO - Loading lsa matrices from dumps...
2024-10-03 14:26:39,324 - cartodata.operations - INFO - Loaded articles matrix of size (100, 4865).
2024-10-03 14:26:39,332 - cartodata.operations - INFO - Loaded authors matrix of size (100, 783).
2024-10-03 14:26:39,338 - cartodata.operations - INFO - Loaded labs matrix of size (100, 677).
2024-10-03 14:26:39,377 - cartodata.operations - INFO - Loaded words matrix of size (100, 5257).
2024-10-03 14:26:39,460 - cartodata.operations - INFO - Loaded articles score vector of length 4865.
2024-10-03 14:26:39,466 - cartodata.operations - INFO - Loaded authors score vector of length 783.
2024-10-03 14:26:39,471 - cartodata.operations - INFO - Loaded labs score vector of length 677.
2024-10-03 14:26:39,485 - cartodata.operations - INFO - Loaded words score vector of length 5257.
2024-10-03 14:26:39,525 - cartodata.operations - INFO - Loaded articles score vector of length 2131.
2024-10-03 14:26:39,530 - cartodata.operations - INFO - Loaded authors score vector of length 396.
2024-10-03 14:26:39,534 - cartodata.operations - INFO - Loaded labs score vector of length 387.
2024-10-03 14:26:39,542 - cartodata.operations - INFO - Loaded words score vector of length 2850.
/usr/local/lib/python3.9/site-packages/umap/umap_.py:1945: UserWarning: n_jobs value 1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1044: RuntimeWarning: overflow encountered in cast
epochs_per_sample[m].astype(np.float32) / negative_sample_rate
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1049: RuntimeWarning: overflow encountered in cast
epoch_of_next_sample.append(epochs_per_sample[m].astype(np.float32))
2024-10-03 14:29:14,632 - cartodata.pipeline.base - INFO - Saving matrices for aligned_umap ...
2024-10-03 14:29:14,635 - cartodata.operations - INFO - Saving articles matrix of size (2, 4865) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/articles_aligned_umap.npy.gz.
2024-10-03 14:29:14,636 - cartodata.operations - INFO - Saving authors matrix of size (2, 783) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/authors_aligned_umap.npy.gz.
2024-10-03 14:29:14,637 - cartodata.operations - INFO - Saving labs matrix of size (2, 677) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/labs_aligned_umap.npy.gz.
2024-10-03 14:29:14,639 - cartodata.operations - INFO - Saving words matrix of size (2, 5257) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/words_aligned_umap.npy.gz.
2024-10-03 14:29:14,640 - cartodata.operations - INFO - Saving articles score vector of length 4865 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/articles_62027e49a8db161bscores.
2024-10-03 14:29:14,707 - cartodata.operations - INFO - Saving authors score vector of length 783 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/authors_62027e49a8db161bscores.
2024-10-03 14:29:14,715 - cartodata.operations - INFO - Saving labs score vector of length 677 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/labs_62027e49a8db161bscores.
2024-10-03 14:29:14,722 - cartodata.operations - INFO - Saving words score vector of length 5257 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/words_62027e49a8db161bscores.
(2, 5257)
(2, 5257)
(2, 5257)
(2, 5257)
2024-10-03 14:29:14,826 - cartodata.pipeline.base - INFO - Entity matrix for nature hl_clusters does not exist for kmeans.
2024-10-03 14:29:14,826 - cartodata.pipeline.base - INFO - Loading mat matrices from dumps...
2024-10-03 14:29:14,830 - cartodata.operations - INFO - Loaded articles matrix of size (4865, 4865).
2024-10-03 14:29:14,835 - cartodata.operations - INFO - Loaded authors matrix of size (4865, 783).
2024-10-03 14:29:14,842 - cartodata.operations - INFO - Loaded labs matrix of size (4865, 677).
2024-10-03 14:29:14,867 - cartodata.operations - INFO - Loaded words matrix of size (4865, 5257).
2024-10-03 14:29:14,957 - cartodata.operations - INFO - Loaded articles score vector of length 4865.
2024-10-03 14:29:14,962 - cartodata.operations - INFO - Loaded authors score vector of length 783.
2024-10-03 14:29:14,966 - cartodata.operations - INFO - Loaded labs score vector of length 677.
2024-10-03 14:29:14,978 - cartodata.operations - INFO - Loaded words score vector of length 5257.
2024-10-03 14:29:14,979 - cartodata.pipeline.base - INFO - Loading lsa matrices from dumps...
2024-10-03 14:29:15,015 - cartodata.operations - INFO - Loaded articles matrix of size (100, 4865).
2024-10-03 14:29:15,022 - cartodata.operations - INFO - Loaded authors matrix of size (100, 783).
2024-10-03 14:29:15,028 - cartodata.operations - INFO - Loaded labs matrix of size (100, 677).
2024-10-03 14:29:15,067 - cartodata.operations - INFO - Loaded words matrix of size (100, 5257).
2024-10-03 14:29:15,068 - cartodata.pipeline.base - INFO - Loading aligned_umap matrices from dumps...
2024-10-03 14:29:15,069 - cartodata.operations - INFO - Loaded articles matrix of size (2, 4865).
2024-10-03 14:29:15,070 - cartodata.operations - INFO - Loaded authors matrix of size (2, 783).
2024-10-03 14:29:15,071 - cartodata.operations - INFO - Loaded labs matrix of size (2, 677).
2024-10-03 14:29:15,072 - cartodata.operations - INFO - Loaded words matrix of size (2, 5257).
2024-10-03 14:29:15,073 - cartodata.pipeline.clustering - INFO - Starting kmeans clustering for hl_clusters. Nb clusters = 8.
TIMEIT 'create_kmeans_clusters' 97.22 ms
2024-10-03 14:29:15,171 - cartodata.pipeline.clustering - INFO - Starting kmeans clustering for ml_clusters. Nb clusters = 24.
TIMEIT 'create_kmeans_clusters' 229.60 ms
2024-10-03 14:29:15,402 - cartodata.operations - INFO - Saving hl_clusters score vector of length 8 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/hl_clusters_scores.
2024-10-03 14:29:15,405 - cartodata.operations - INFO - Saving ml_clusters score vector of length 24 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/ml_clusters_scores.
2024-10-03 14:29:15,407 - cartodata.pipeline.base - INFO - Saving matrices for lsa ...
2024-10-03 14:29:15,408 - cartodata.operations - INFO - Saving hl_clusters matrix of size (100, 8) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/hl_clusters_lsa.npy.gz.
2024-10-03 14:29:15,409 - cartodata.operations - INFO - Saving ml_clusters matrix of size (100, 24) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/ml_clusters_lsa.npy.gz.
2024-10-03 14:29:15,409 - cartodata.pipeline.base - INFO - Saving matrices for aligned_umap ...
2024-10-03 14:29:15,410 - cartodata.operations - INFO - Saving hl_clusters matrix of size (2, 8) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/hl_clusters_aligned_umap.npy.gz.
2024-10-03 14:29:15,410 - cartodata.operations - INFO - Saving ml_clusters matrix of size (2, 24) into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/ml_clusters_aligned_umap.npy.gz.
2024-10-03 14:29:15,410 - cartodata.operations - INFO - Saving hl_clusters cluster labels into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/hl_clusters_labels.json
2024-10-03 14:29:15,443 - cartodata.operations - INFO - Saving ml_clusters cluster labels into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/ml_clusters_labels.json
2024-10-03 14:29:15,510 - cartodata.operations - INFO - Saving hl_clusters score vector of length 8 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/hl_clusters_eval_posscores.
2024-10-03 14:29:15,513 - cartodata.operations - INFO - Saving ml_clusters score vector of length 24 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/ml_clusters_eval_posscores.
2024-10-03 14:29:15,516 - cartodata.operations - INFO - Saving hl_clusters score vector of length 8 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/hl_clusters_eval_negscores.
2024-10-03 14:29:15,518 - cartodata.operations - INFO - Saving ml_clusters score vector of length 24 into /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/ml_clusters_eval_negscores.
2024-10-03 14:29:15,572 - cartodata.pipeline.base - INFO - Loading aligned_umap matrices from dumps...
2024-10-03 14:29:15,574 - cartodata.operations - INFO - Loaded articles matrix of size (2, 4865).
2024-10-03 14:29:15,575 - cartodata.operations - INFO - Loaded authors matrix of size (2, 783).
2024-10-03 14:29:15,576 - cartodata.operations - INFO - Loaded labs matrix of size (2, 677).
2024-10-03 14:29:15,578 - cartodata.operations - INFO - Loaded words matrix of size (2, 5257).
2024-10-03 14:29:15,903 - cartodata.operations - INFO - Loaded hl_clusters score vector of length 8.
2024-10-03 14:29:15,906 - cartodata.operations - INFO - Loaded ml_clusters score vector of length 24.
2024-10-03 14:29:15,907 - cartodata.pipeline.base - INFO - Loading lsa matrices from dumps...
2024-10-03 14:29:15,908 - cartodata.operations - INFO - Loaded hl_clusters matrix of size (100, 8).
2024-10-03 14:29:15,909 - cartodata.operations - INFO - Loaded ml_clusters matrix of size (100, 24).
2024-10-03 14:29:15,909 - cartodata.pipeline.base - INFO - Loading aligned_umap matrices from dumps...
2024-10-03 14:29:15,910 - cartodata.operations - INFO - Loaded hl_clusters matrix of size (2, 8).
2024-10-03 14:29:15,911 - cartodata.operations - INFO - Loaded ml_clusters matrix of size (2, 24).
2024-10-03 14:29:15,916 - cartodata.operations - INFO - Loaded hl_clusters score vector of length 8.
2024-10-03 14:29:15,919 - cartodata.operations - INFO - Loaded ml_clusters score vector of length 24.
2024-10-03 14:29:15,923 - cartodata.operations - INFO - Loaded hl_clusters score vector of length 8.
2024-10-03 14:29:15,926 - cartodata.operations - INFO - Loaded ml_clusters score vector of length 24.
2024-10-03 14:29:15,927 - cartodata.operations - INFO - Loaded hl_clusters cluster labels from file /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/hl_clusters_labels.json.
2024-10-03 14:29:15,933 - cartodata.operations - INFO - Loaded ml_clusters cluster labels from file /builds/2mk6rsew/0/hgozukan/cartolabe-data/dumps/lisn/3.2.0/s1/ml_clusters_labels.json.
<IPython.core.display.Image object>
Total running time of the script: (10 minutes 50.528 seconds)