Note
Go to the end to download the full example code.
Extracting and processing LISN data for Cartolabe with Aligned UMAP¶
( Doc2vec projection)
In this example we will:
extract entities (authors, articles, labs, words) from a collection of scientific articles
project those entities in 2 dimensions using aligned UMAP
cluster them
find their nearest neighbors.
This example uses 6 datasets containing articles from HAL (https://hal.archives-ouvertes.fr/) published by authors from LISN (Laboratoire Interdisciplinaire des Sciences du Numérique). Each dataset contains cumulative data starting from 2010-2012 to 2010-2022 (inclusive).
Using aligned UMAP will help us align UMAP embeddings for consecutive datasets.
# %matplotlib widget
Download data¶
We will start by downloading 6 datasets from HAL.
from cartodata.scraping import scrape_hal, process_domain_column # noqa
import os # noqa
def fetch_data(from_year, to_year, struct_ids, struct='hal'):
"""
Fetch scientific publications of struct from HAL.
"""
filename = f"../datas/{struct.lower()}_{from_year}_{to_year - 1}.csv"
if os.path.exists(filename):
return
filters = {}
if struct:
filters['structId_i'] = struct_ids
years = range(from_year, to_year)
df = scrape_hal(struct, filters, years, cool_down=2)
process_domain_column(df)
# Save the dataframe into a csv file
df.to_csv(filename)
return df
# Fetch LRI data from year 2010 to 2022 inclusive
for i in range(2, 14, 2):
fetch_data(2010, 2011+i, "(1050003 2544)", 'lisn')
/builds/2mk6rsew/0/hgozukan/cartolabe-data/cartodata/scraping.py:116: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation.
df = pd.concat([df, year_df])
/builds/2mk6rsew/0/hgozukan/cartolabe-data/cartodata/scraping.py:116: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation.
df = pd.concat([df, year_df])
/builds/2mk6rsew/0/hgozukan/cartolabe-data/cartodata/scraping.py:116: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation.
df = pd.concat([df, year_df])
/builds/2mk6rsew/0/hgozukan/cartolabe-data/cartodata/scraping.py:116: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation.
df = pd.concat([df, year_df])
/builds/2mk6rsew/0/hgozukan/cartolabe-data/cartodata/scraping.py:116: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation.
df = pd.concat([df, year_df])
/builds/2mk6rsew/0/hgozukan/cartolabe-data/cartodata/scraping.py:116: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated. In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes. To retain the old behavior, exclude the relevant entries before the concat operation.
df = pd.concat([df, year_df])
Load data¶
Now we will load each downloaded dataset to a dataframe.
import pandas as pd # noqa
def load_data(from_year, total, struct):
dataframes = []
for i in range(total):
file_name = f"../datas/{struct.lower()}_{from_year}_{from_year + ( 2 + 2 * i)}.csv"
print(file_name)
df = pd.read_csv(file_name, index_col=0)
dataframes.append(df)
return dataframes
dataframes = load_data(2010, 6, "lisn")
dataframes[0].head()
../datas/lisn_2010_2012.csv
../datas/lisn_2010_2014.csv
../datas/lisn_2010_2016.csv
../datas/lisn_2010_2018.csv
../datas/lisn_2010_2020.csv
../datas/lisn_2010_2022.csv
We can list the total number of articles starting from 2010 to a certain year.
for i in range(6):
print(f"{2010 + (i*2) + 2} => {dataframes[i].shape[0]}")
2012 => 765
2014 => 1361
2016 => 1983
2018 => 2514
2020 => 2945
2022 => 2987
Creating correspondance matrices for each entity type¶
From this table of articles, we want to extract matrices that will map the correspondance between these articles and the entities we want to use.
from cartodata.loading import load_comma_separated_column # noqa
from cartodata.loading import load_identity_column # noqa
def create_correspondance_matrices(func, dataframes, column_name):
matrices = []
scores = []
for i in range(len(dataframes)):
mat, score = func(dataframes[i], column_name)
matrices.append(mat)
scores.append(score)
return matrices, scores
The load_comma_separated_column function takes in a dataframe and the name of a column and returns two objects:
a sparse matrix a pandas Series
Each column of the sparse matrix, corresponds to the entity specified by column_name and each row corresponds to an article.
Labs¶
Similarly, we can create matrices for the labs by simply passing the structAcronym_s column to the function.
matrices_labs, scores_labs = create_correspondance_matrices(
load_comma_separated_column, dataframes, 'structAcronym_s')
We can see the number of (articles, labs) for each year:
for i in range(len(matrices_labs)):
print(f"{2010 + (i*2) + 2} => {matrices_labs[i].shape}")
2012 => (765, 602)
2014 => (1361, 838)
2016 => (1983, 1041)
2018 => (2514, 1254)
2020 => (2945, 1426)
2022 => (2987, 1447)
Let’s have a look at the number of articles per lab for each year:
df_scores_labs = pd.concat(scores_labs, axis=1).reset_index().rename(
columns={i: f"{2010 + (i*2) + 2}" for i in range(10)}
)
df_scores_labs.head(20)
""
df_scores_labs.describe()
Filtering low score entities¶
A lot of the authors and labs that we just extracted from the dataframe have a very low score, which means they’re only linked to one or two articles. To improve the quality of our data, we’ll filter the entities by removing those that appear less than certain number of times.
from cartodata.operations import filter_min_score # noqa
def filter_low_score(matrices, scores, count, entity):
filtered_matrices = []
filtered_scores = []
for i, (matrix, score) in enumerate(zip(matrices, scores)):
before = len(score)
filtered_mat, filtered_score = filter_min_score(matrix, score, count)
filtered_matrices.append(filtered_mat)
filtered_scores.append(filtered_score)
print(f'Removed {before - len(filtered_score)} {entity}s with less than ' +
f'{count} articles from a total of {before} {entity}s.')
print(f'Working with {len(filtered_score)} {entity} for year {2010 + (i*2) + 2}.\n')
return filtered_matrices, filtered_scores
Authors¶
We will remove the authors with less than 5 publications.
auth_pub_count = 5
filtered_auth_matrices, filtered_auth_scores = filter_low_score(
matrices_auth, scores_auth, auth_pub_count, "author")
Removed 1578 authors with less than 5 articles from a total of 1666 authors.
Working with 88 author for year 2012.
Removed 2179 authors with less than 5 articles from a total of 2338 authors.
Working with 159 author for year 2014.
Removed 2887 authors with less than 5 articles from a total of 3135 authors.
Working with 248 author for year 2016.
Removed 3566 authors with less than 5 articles from a total of 3882 authors.
Working with 316 author for year 2018.
Removed 4168 authors with less than 5 articles from a total of 4554 authors.
Working with 386 author for year 2020.
Removed 4258 authors with less than 5 articles from a total of 4652 authors.
Working with 394 author for year 2022.
Labs¶
We will remove the labs with less than 20 publications.
lab_pub_count = 20
filtered_lab_matrices, filtered_lab_scores = filter_low_score(
matrices_labs, scores_labs, lab_pub_count, "labs")
Removed 566 labss with less than 20 articles from a total of 602 labss.
Working with 36 labs for year 2012.
Removed 762 labss with less than 20 articles from a total of 838 labss.
Working with 76 labs for year 2014.
Removed 938 labss with less than 20 articles from a total of 1041 labss.
Working with 103 labs for year 2016.
Removed 1131 labss with less than 20 articles from a total of 1254 labss.
Working with 123 labs for year 2018.
Removed 1286 labss with less than 20 articles from a total of 1426 labss.
Working with 140 labs for year 2020.
Removed 1305 labss with less than 20 articles from a total of 1447 labss.
Working with 142 labs for year 2022.
Words¶
For the words, it’s a bit trickier because we want to extract n-grams (groups of n terms) instead of just comma separated values. We’ll call the load_text_column which uses scikit-learn’s CountVectorizer <https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html>_ to create a vocabulary and map the tokens.
from cartodata.loading import load_text_column # noqa
from sklearn.feature_extraction import text as sktxt # noqa
from cartodata.operations import normalize_tfidf # noqa
def process_words(dataframes, stopwords):
words_matrices = []
words_scores = []
for df in dataframes:
df['text'] = df['en_abstract_s'] + ' ' \
+ df['en_keyword_s'].astype(str) + ' ' \
+ df['en_title_s'].astype(str) + ' ' \
+ df['en_domainAllCodeLabel_fs'].astype(str)
words_mat, words_score = load_text_column(df['text'],
4,
10,
0.05,
stopwords=stopwords)
# apply term-frequency times inverse document-frequency normalization
words_mat = normalize_tfidf(words_mat)
words_matrices.append(words_mat)
words_scores.append(words_score)
return words_matrices, words_scores
with open('../datas/stopwords.txt', 'r') as stop_file:
stopwords = sktxt.ENGLISH_STOP_WORDS.union(
set(stop_file.read().splitlines()))
words_matrices, words_scores = process_words(dataframes, stopwords)
We can list number of terms per article for each year:
for i in range(len(words_matrices)):
print(f"{2010 + (i*2) + 2} => {words_matrices[i].shape}")
2012 => (765, 1015)
2014 => (1361, 1801)
2016 => (1983, 2483)
2018 => (2514, 3045)
2020 => (2945, 3512)
2022 => (2987, 3560)
Articles¶
Finally, we need to create a matrix that simply maps each article to itself.
matrices_article, scores_article = create_correspondance_matrices(
load_identity_column, dataframes, 'en_title_s')
""
for i in range(len(matrices_article)):
print(f"{2010 + (i*2) + 2} => {matrices_article[i].shape}")
2012 => (765, 765)
2014 => (1361, 1361)
2016 => (1983, 1983)
2018 => (2514, 2514)
2020 => (2945, 2945)
2022 => (2987, 2987)
Dimension reduction¶
One way to see the matrices that we created is as coordinates in the space of all articles. What we want to do is to reduce the dimension of this space to make it easier to work with and see.
Doc2vec projection¶
This example uses the Doc2vec technique to identify keywords in our data and thus reduce the number of rows in our matrices. The doc2vec_projection method takes at least four arguments:
the number of dimensions you want to keep
the matrix identifier of documents/words frequency
a list of matrices to project
the list of text to index (it is possible to add doc2vec parameters with
the same syntax of the gensim doc2vec function).
It returns a list of matrices projected in the latent space.
We also apply an l2 normalization to each feature of the projected matrices.
from cartodata.projection import doc2vec_projection # noqa
from cartodata.operations import normalize_l2 # noqa
dimensions = [20, 20, 30, 30, 40, 40]
def run_doc2vec_projection(articles_matrices, auth_matrices, words_matrices,
lab_matrices, dimensions, dataframes, column_name):
doc2vec_matrices = []
for articles_mat, auth_mat, words_mat, labs_mat, dim, df in zip(
articles_matrices, auth_matrices, words_matrices, lab_matrices, dimensions, dataframes):
doc2vec_mat = doc2vec_projection(dim,
3,
[articles_mat, auth_mat, words_mat, labs_mat],
df[column_name])
doc2vec_mat = list(map(normalize_l2, doc2vec_mat))
doc2vec_matrices.append(doc2vec_mat)
return doc2vec_matrices
doc2vec_matrices = run_doc2vec_projection(matrices_article, filtered_auth_matrices,
words_matrices, filtered_lab_matrices,
dimensions, dataframes, 'text')
List (number_of_entity, dimensions) per year:
for j, type_ in enumerate(["ARTICLES", "AUTHORS", "WORDS", "LABS"]):
print(type_)
for i in range(len(doc2vec_matrices)):
print(f"201{i} => {doc2vec_matrices[i][j].shape}")
ARTICLES
2010 => (20, 765)
2011 => (20, 1361)
2012 => (30, 1983)
2013 => (30, 2514)
2014 => (40, 2945)
2015 => (40, 2987)
AUTHORS
2010 => (20, 88)
2011 => (20, 159)
2012 => (30, 248)
2013 => (30, 316)
2014 => (40, 386)
2015 => (40, 394)
WORDS
2010 => (20, 1015)
2011 => (20, 1801)
2012 => (30, 2483)
2013 => (30, 3045)
2014 => (40, 3512)
2015 => (40, 3560)
LABS
2010 => (20, 36)
2011 => (20, 76)
2012 => (30, 103)
2013 => (30, 123)
2014 => (40, 140)
2015 => (40, 142)
Aligned UMAP projection¶
To use aligned UMAP, we need to create relations between each consecutive dataset that maps each entity index (article, author, word, lab) in one dataset to corresponding entity index in the following dataset.
We will also create color mappings to view each entity in consecutive maps with the same color.
import numpy as np # noqa
def make_relation(from_df, to_df):
# create a new dataframe with index from from_df, and values as integers
# starting from 0 to the length of from_df
left = pd.DataFrame(data=np.arange(len(from_df)), index=from_df.index)
# create a new dataframe with index from to_df, and values as integers
# starting from 0 to the length of to_df
right = pd.DataFrame(data=np.arange(len(to_df)), index=to_df.index)
# merge left and right dataframes on the intersection of keys of both
# dataframes preserving the order of left keys
merge = pd.merge(left, right, how="inner",
left_index=True, right_index=True)
return dict(merge.values)
def generate_relations(filtered_scores):
relations = []
for i in range(len(filtered_scores) - 1):
relation = make_relation(filtered_scores[i], filtered_scores[i+1])
relations.append(relation)
return relations
def generate_colors(filtered_scores):
colors = []
for i in range(len(filtered_scores)):
color = make_relation(filtered_scores[i], filtered_scores[-1])
colors.append(color)
return colors
def concat_scores(scores_article, scores_auth, scores_word, scores_lab):
"""Concatenates article, auth, words and labs score for each year and
returns the concatenated frames in a list.
"""
concatenated_scores = []
for score_article, score_auth, score_word, score_lab in zip(
scores_article, scores_auth, scores_word, scores_lab
):
concatenated_scores.append(
pd.concat([score_article, score_auth, score_word, score_lab]))
return concatenated_scores
filtered_scores = concat_scores(
scores_article, filtered_auth_scores, words_scores, filtered_lab_scores)
relations = generate_relations(filtered_scores)
colors_auth = generate_colors(filtered_auth_scores)
colors_words = generate_colors(words_scores)
colors_lab = generate_colors(filtered_lab_scores)
colors_articles = generate_colors(scores_article)
We should get transpose of each doc2vec matrix to be able to process by UMAP.
def get_transpose(doc2vec_matrices):
transposed_mats = []
for doc2vec in doc2vec_matrices:
transposed_mats.append(np.hstack(doc2vec).T)
return transposed_mats
transposed_mats = get_transpose(doc2vec_matrices)
We create an AlignedUMAP instance to generate 10 maps for the 10 datasets.
from umap.aligned_umap import AlignedUMAP # noqa
n_neighbors = [10, 10, 20, 20, 50, 80]
min_dists = [0.05, 0.01, 0.01, 0.01, 0.001, 0.001]
spreads = [1, 1, 1, 1, 1, 1]
n = 6
reducer = AlignedUMAP(
n_neighbors=n_neighbors[:n],
min_dist=min_dists[:n],
# n_neighbors=10,
# min_dist=0.05,
init='random',
n_epochs=300)
Aligned UMAP requires at least two datasets and the mapping between the entities of the datasets. We can change the number of initial datasets changing the value of the variable n.
reducer.fit_transform(transposed_mats[:n], relations=relations[:n-1])
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1044: RuntimeWarning: overflow encountered in cast
epochs_per_sample[m].astype(np.float32) / negative_sample_rate
/usr/local/lib/python3.9/site-packages/umap/layouts.py:1049: RuntimeWarning: overflow encountered in cast
epoch_of_next_sample.append(epochs_per_sample[m].astype(np.float32))
ListType[array(float32, 2d, C)]([[[ 1.636822 4.3104906]
[-2.5236168 -2.0890348]
[ 4.760189 -5.08321 ]
...
[ 6.92317 -1.5047461]
[ 6.876193 -1.5049655]
[ 5.191547 -0.0401044]], [[-1.5608968 -2.107365 ]
[-2.3126545 -2.7574205]
[ 4.386353 2.9121335]
...
[ 4.935302 -1.7496797]
[ 3.0446644 0.941432 ]
[ 3.0300503 1.4598998]], [[-1.9637237 -4.284528 ]
[-1.5079978 -4.3302937 ]
[ 6.923994 1.4484942 ]
...
[ 0.76254576 -4.921924 ]
[ 1.286736 -0.81693447]
[ 1.5955446 -0.95411617]], [[-1.6925725 -4.697059 ]
[-0.98491496 -3.441007 ]
[ 6.1821823 0.7781479 ]
...
[ 3.5565672 -2.5444643 ]
[ 7.748843 0.28574288]
[ 3.465931 -0.65967333]], [[-1.3731371 -4.6526 ]
[-1.0955343 -4.0270705 ]
[ 3.599194 -1.14507 ]
...
[ 2.9202418 -0.47803032]
[ 5.5627213 -1.3473547 ]
[ 2.0311272 -1.689975 ]], [[-0.7290484 -4.4938498 ]
[-0.9665915 -4.021041 ]
[ 3.1820416 -0.44369522]
...
[ 5.6325536 -1.2587452 ]
[ 2.156493 -1.4376152 ]
[ 5.267645 -0.9021303 ]]])
Then we will generate maps for the remaining dataset one by one by feeding the reducer with the corresponding matrix and relation dictionary.
def update_reducer(reducer, matrices, relations,
n_neighbors, min_dists, spreads):
for mat, rel, n_neighbor, min_dist, spread in zip(
matrices, relations, n_neighbors, min_dists, spreads
):
# reducer.update(mat, relations=rel)
reducer.update(mat, relations=rel, n_neighbors=n_neighbor,
min_dist=min_dist, spread=spread, verbose=True)
update_reducer(reducer, transposed_mats[n:], relations[n-1:],
n_neighbors[n:], min_dists[n:], spreads[n:])
""
for embedding in reducer.embeddings_:
print(embedding.shape)
(1904, 2)
(3397, 2)
(4817, 2)
(5998, 2)
(6983, 2)
(7083, 2)
Plot maps¶
To plot the maps, we will reorganize the data.
We will add author data to embeddings, so that we can visualize author name when the plot is hovered.
def update_embeddings_with_entities(embeddings, filtered_scores):
r_embeddings = []
for embedding, score in zip(embeddings, filtered_scores):
score_indices = np.matrix(score.index).T
embd = np.hstack((embedding, score_indices)).T
r_embeddings.append(np.asarray(embd))
return r_embeddings
def decompose_embeddings(embeddings):
article_embeddings = []
auth_embeddings = []
word_embeddings = []
lab_embeddings = []
for i, embedding in enumerate(embeddings):
len_article = len(scores_article[i])
len_auth = len(filtered_auth_scores[i])
len_word = len(words_scores[i])
article_embeddings.append(embeddings[i][:len_article])
auth_embeddings.append(
embeddings[i][len_article: len_article + len_auth])
word_embeddings.append(
embeddings[i][len_article + len_auth:len_article + len_auth + len_word])
lab_embeddings.append(
embeddings[i][len_article + len_auth + len_word:])
return article_embeddings, auth_embeddings, word_embeddings, lab_embeddings
article_embeddings, auth_embeddings, word_embeddings, lab_embeddings = decompose_embeddings(
reducer.embeddings_)
We will add values to embeddings, so that we can visualize article title, author name, word or lab name when the entity is hovered on the plot.
article_embeddings_names = update_embeddings_with_entities(
article_embeddings, scores_article
)
auth_embeddings_names = update_embeddings_with_entities(
auth_embeddings, filtered_auth_scores
)
word_embeddings_names = update_embeddings_with_entities(
word_embeddings, words_scores
)
lab_embeddings_names = update_embeddings_with_entities(
lab_embeddings, filtered_lab_scores
)
We will create a colormap that will map each entity (article, author, word, lab) to same color in every plot.
from matplotlib import cm # noqa
def create_color_map(max_list):
cmap = cm.get_cmap('gist_ncar', len(max_list))
return cmap
cmap_article = create_color_map(scores_article[-1])
cmap_auth = create_color_map(filtered_auth_scores[-1])
cmap_word = create_color_map(words_scores[-1])
cmap_lab = create_color_map(filtered_lab_scores[-1])
/builds/2mk6rsew/0/hgozukan/cartolabe-data/examples/workflow_aligned_lisn_doc2vec_kmeans.py:577: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
cmap = cm.get_cmap('gist_ncar', len(max_list))
Now we can plot all the maps.
import matplotlib.pyplot as plt # noqa
import numpy as np # noqa
import mplcursors # noqa
plt.close('all')
def axis_bounds(embedding):
left, right = embedding.T[0].min(), embedding.T[0].max()
bottom, top = embedding.T[1].min(), embedding.T[1].max()
adj_h, adj_v = (right - left) * 0.1, (top - bottom) * 0.1
return [left - adj_h, right + adj_h, bottom - adj_v, top + adj_v]
ax_bound = axis_bounds(np.vstack(reducer.embeddings_))
ax_bound
def plot(title, embedding_article, embedding_auth, embedding_word, embedding_lab,
colors_article, colors_auth, colors_word, colors_lab,
c_scores=None, c_umap=None, n_clusters=None):
colors_article_list = list(colors_article.values())
colors_auth_list = list(colors_auth.values())
colors_word_list = list(colors_word.values())
colors_lab_list = list(colors_lab.values())
fig, ax = plt.subplots(1, 1, figsize=(10, 9))
ax.set_title(title)
ax.axis(ax_bound)
article = ax.scatter(embedding_article[0], embedding_article[1], c=colors_article_list, cmap=cmap_article,
vmin=0, vmax=len(scores_article[-1]), marker='x', label="article")
auth = ax.scatter(embedding_auth[0], embedding_auth[1], c=colors_auth_list, cmap=cmap_auth,
vmin=0, vmax=len(filtered_auth_scores[-1]), label="auth")
word = ax.scatter(embedding_word[0], embedding_word[1],
vmin=0, vmax=len(words_scores[-1]), marker='+', label="word")
lab = ax.scatter(embedding_lab[0], embedding_lab[1], c=colors_lab_list, cmap=cmap_lab,
vmin=0, vmax=len(filtered_lab_scores[-1]), marker='s', label="lab")
# from https://matplotlib.org/stable/gallery/event_handling/legend_picking.html
leg = ax.legend((article, auth, word, lab), ("article",
"auth", "words", "labs"), fancybox=True, shadow=True)
if n_clusters is not None:
for i in range(n_clusters):
ax.annotate(c_scores.index[i], (c_umap[0, i], c_umap[1, i]))
# crs_article = mplcursors.cursor(article, hover=True)
# @crs_article.connect("add")
# def on_add(sel):
# sel.annotation.set(text=embedding_article[2][sel.target.index])
crs_auth = mplcursors.cursor(auth, hover=True)
@crs_auth.connect("add")
def on_add(sel):
sel.annotation.set(text=embedding_auth[2][sel.target.index])
# crs_word = mplcursors.cursor(word, hover=True)
# @crs_word.connect("add")
# def on_add(sel):
# sel.annotation.set(text=embedding_word[2][sel.target.index])
crs_lab = mplcursors.cursor(lab, hover=True)
@crs_lab.connect("add")
def on_add(sel):
sel.annotation.set(text=embedding_lab[2][sel.target.index])
plt.show()
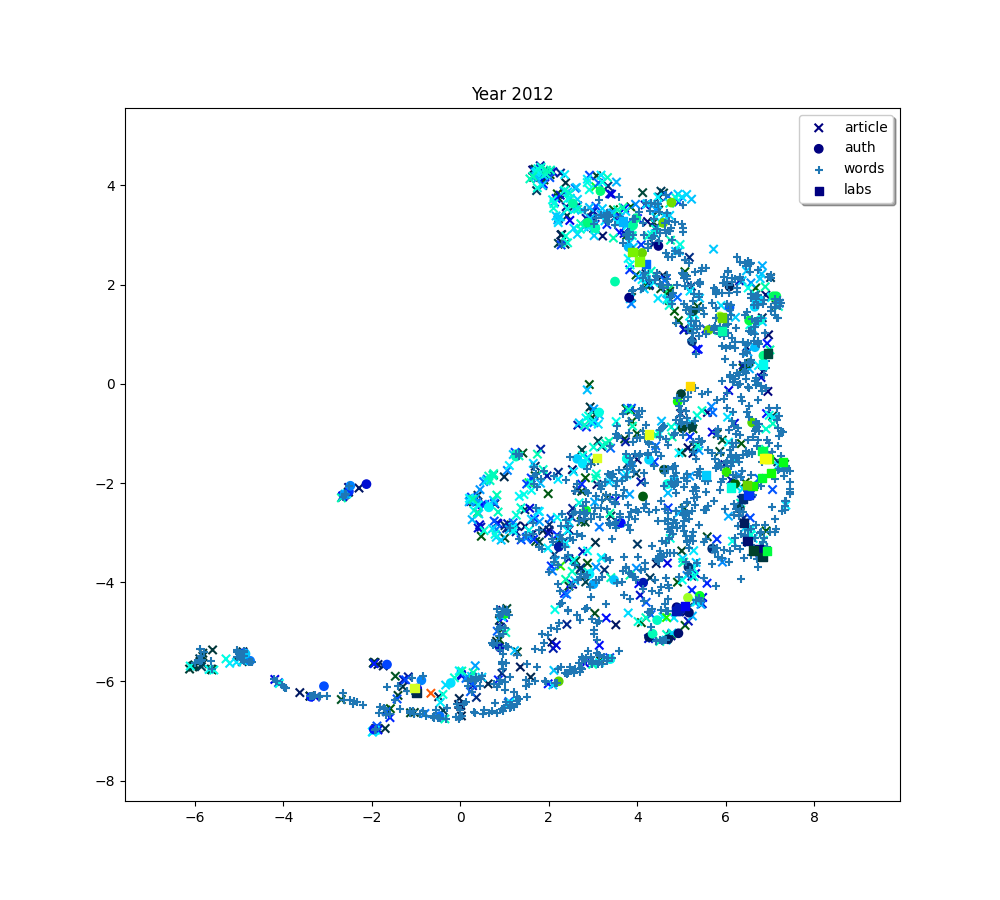
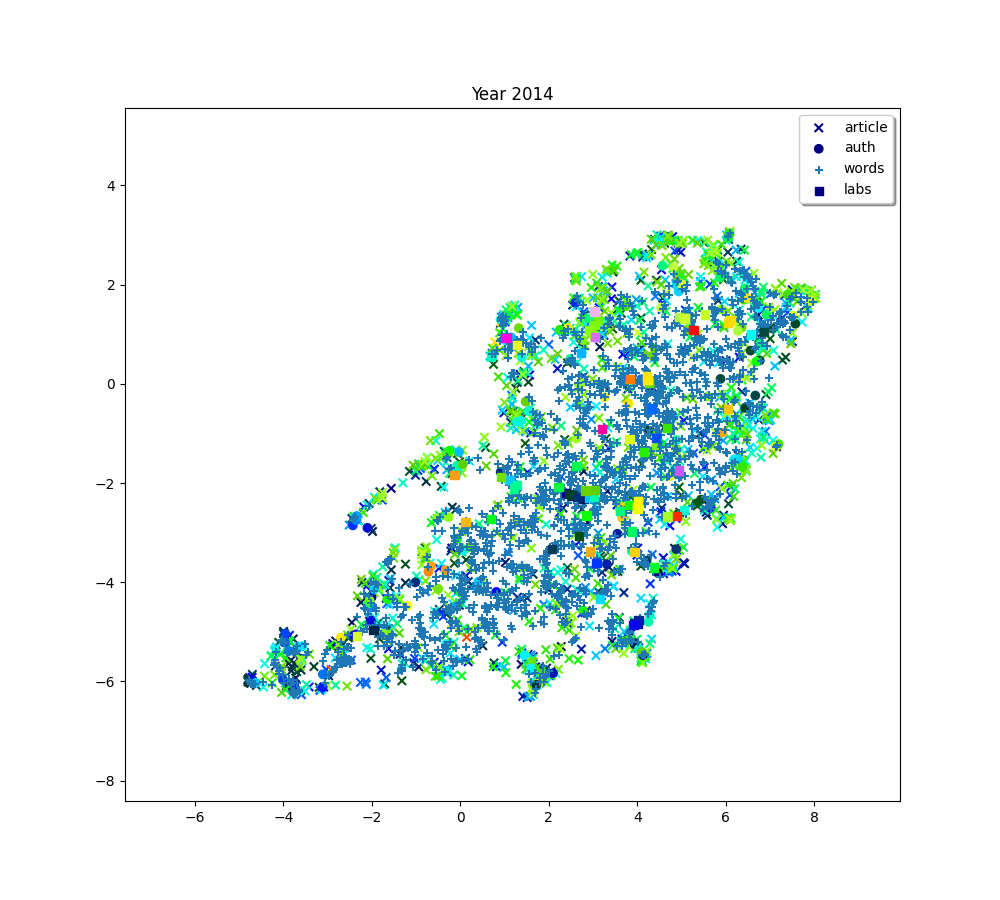
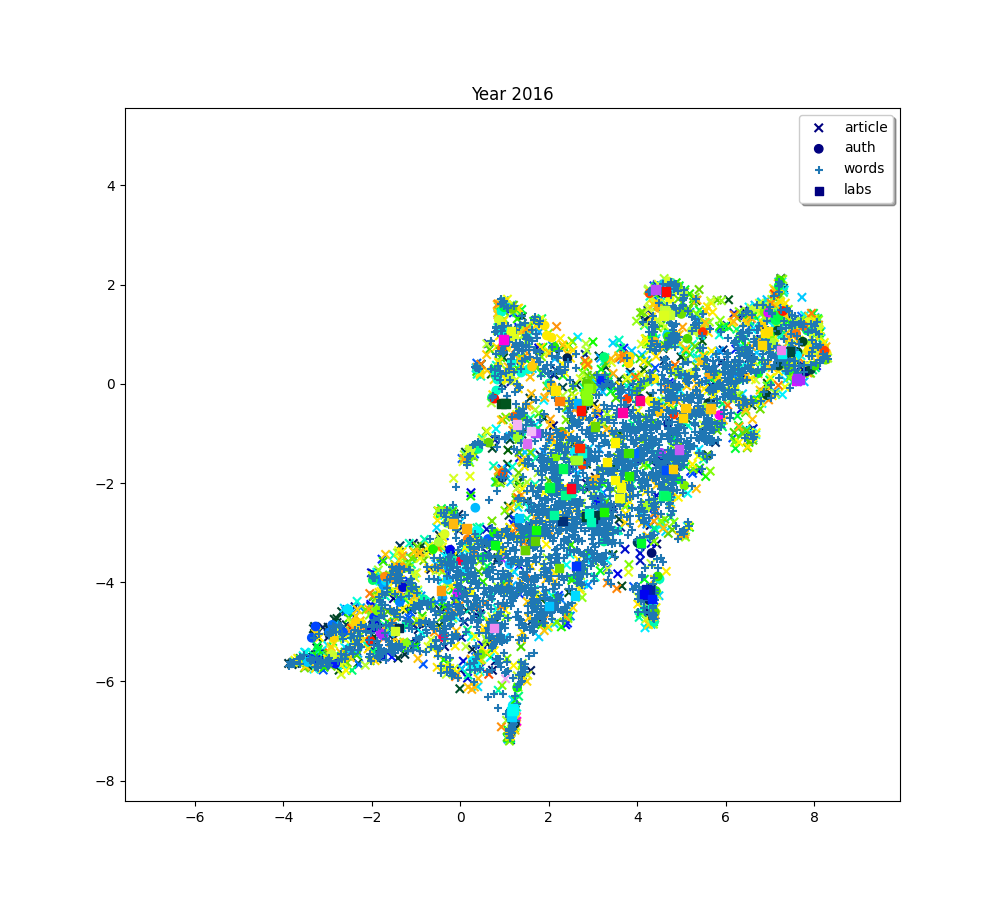
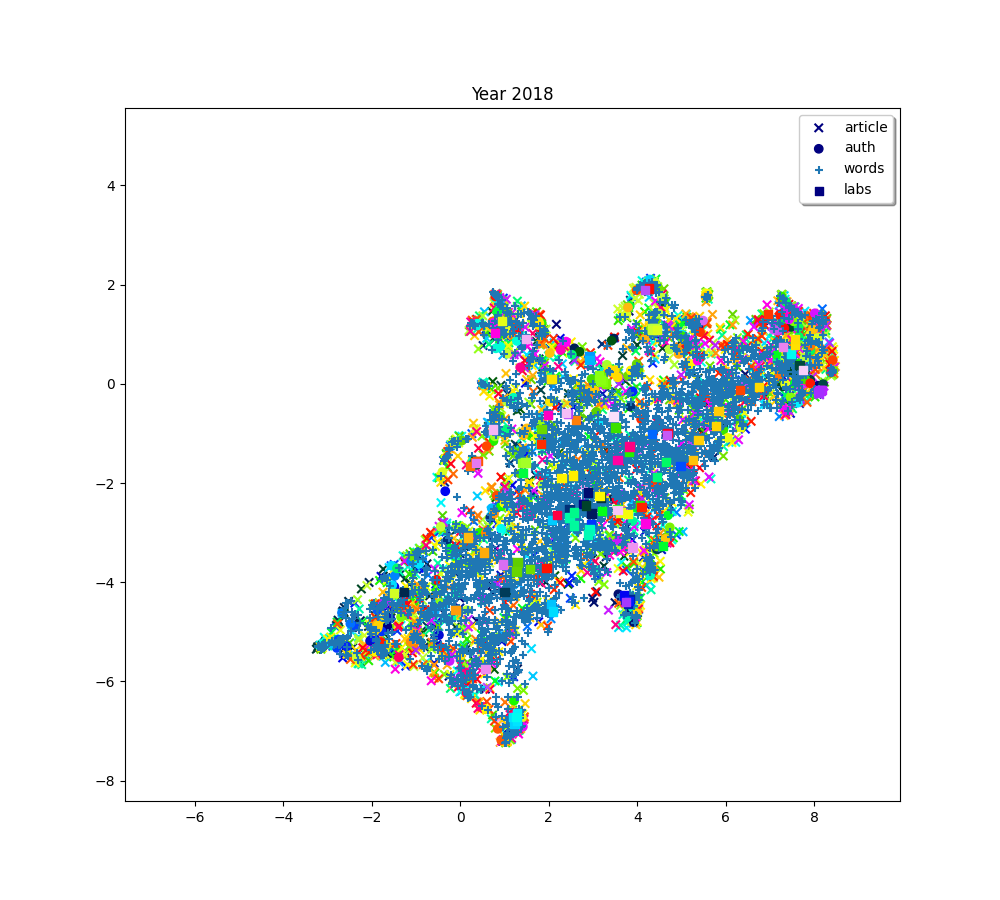
for i, (embedding_article, embedding_auth, embedding_word, embedding_lab,
color_article, color_auth, color_word, color_lab) in enumerate(zip(
article_embeddings_names, auth_embeddings_names, word_embeddings_names, lab_embeddings_names,
colors_articles, colors_auth, colors_words, colors_lab)):
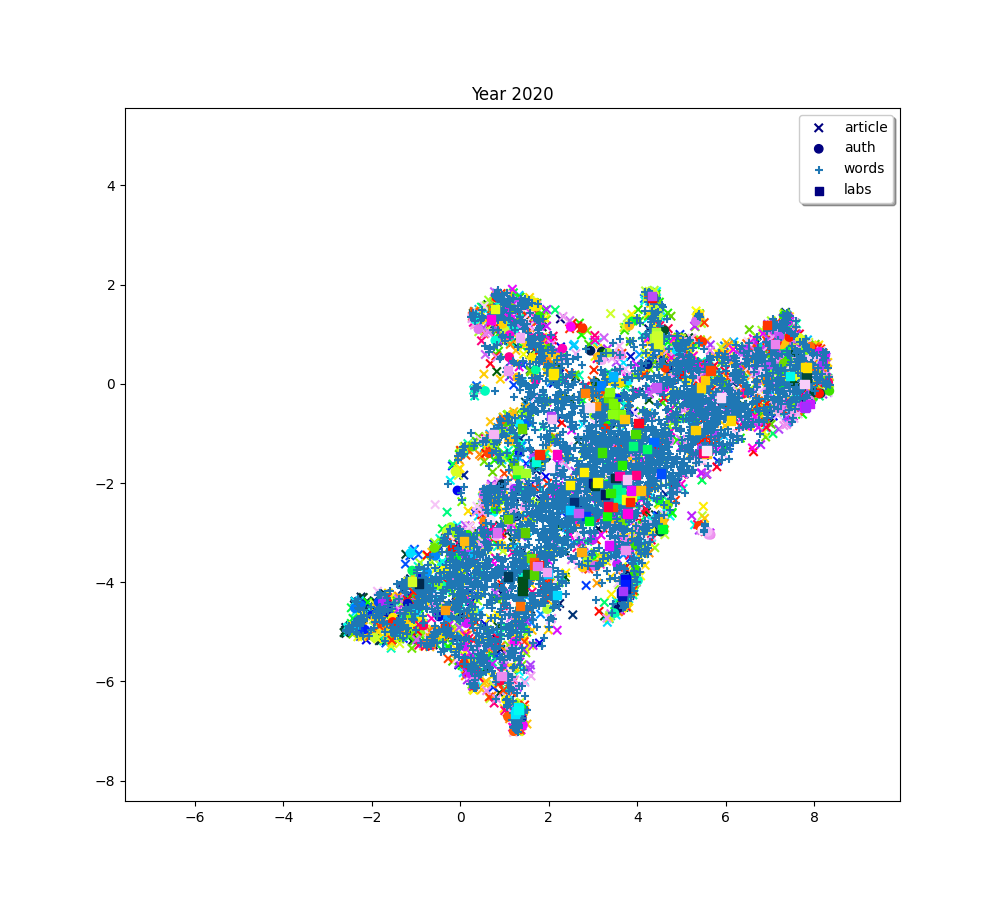
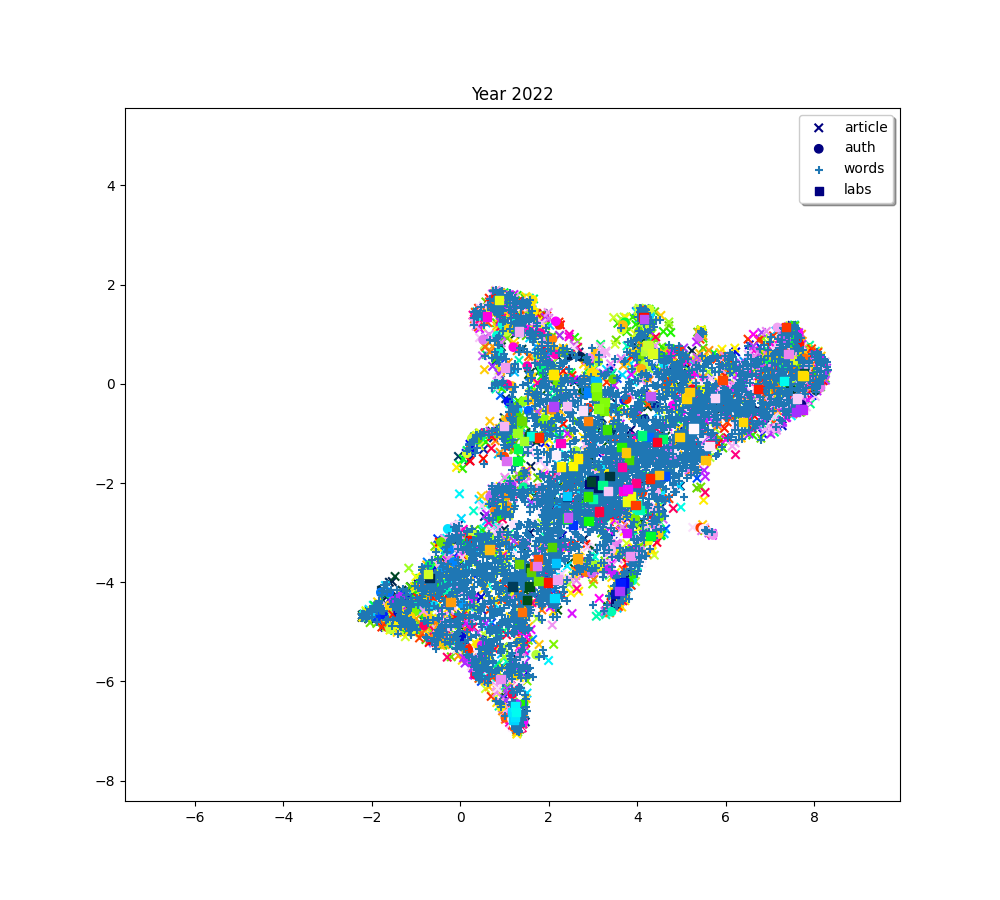
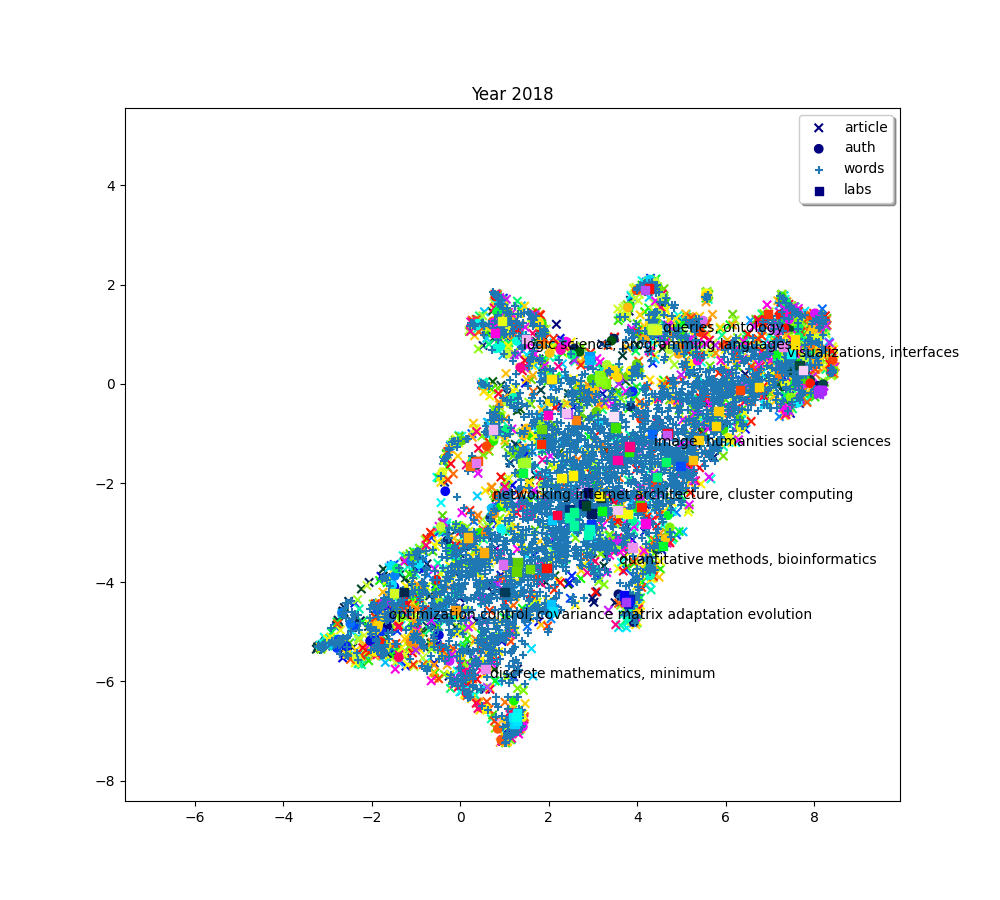
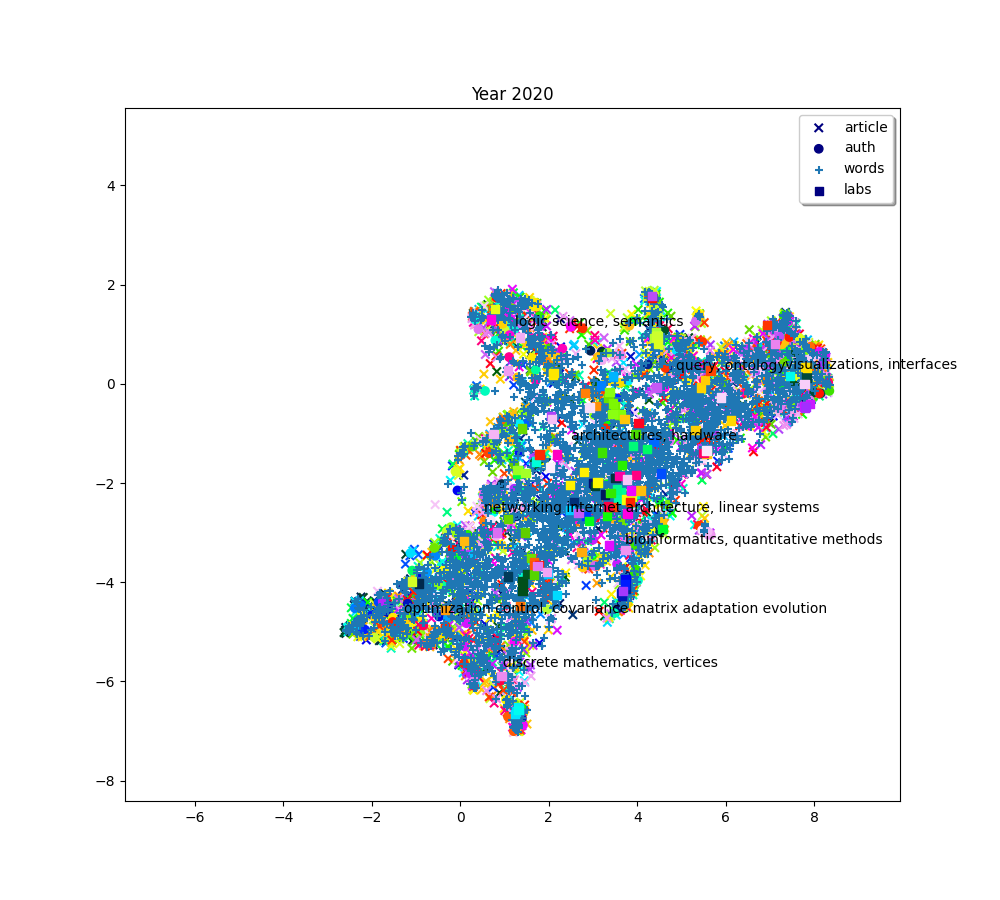
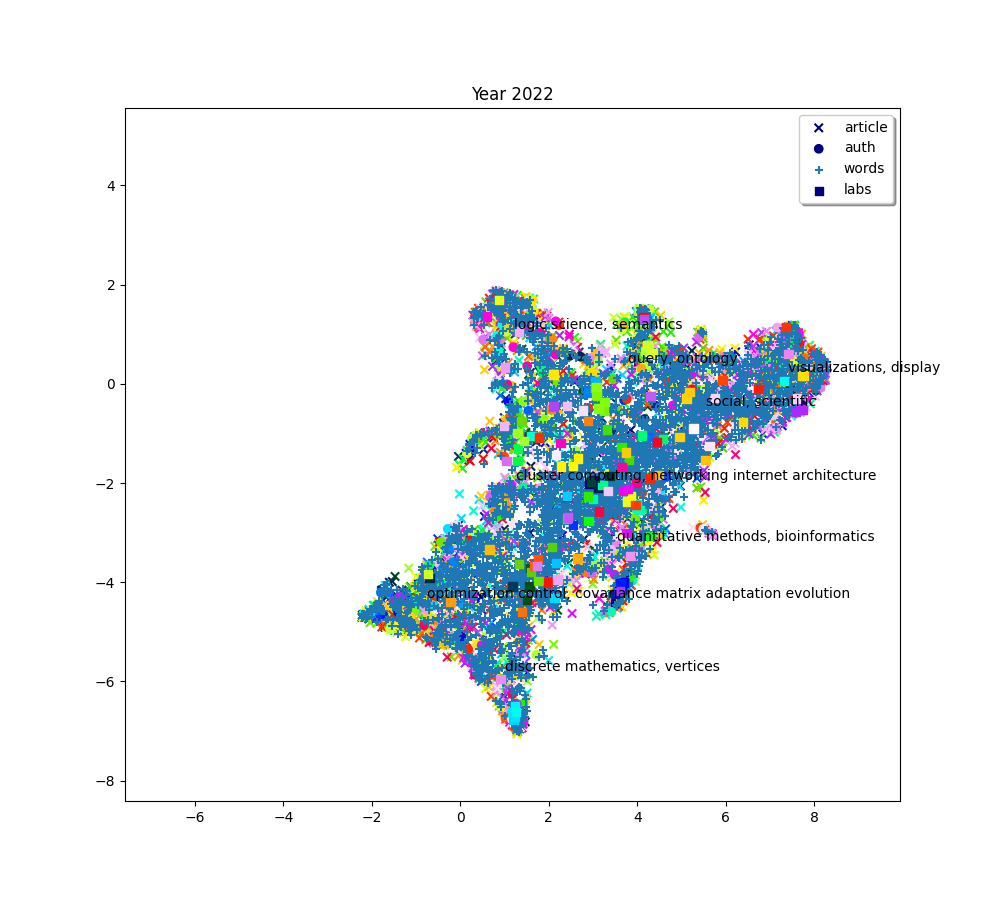
plot(f"Year {2010 + (i * 2) + 2}", embedding_article, embedding_auth, embedding_word, embedding_lab,
color_article, color_auth, color_word, color_lab)
/usr/local/lib/python3.9/site-packages/mplcursors/_pick_info.py:55: UserWarning: No data for colormapping provided via 'c'. Parameters 'vmin', 'vmax' will be ignored
paths = scatter.__wrapped__(*args, **kwargs)
Clustering¶
In order to identify clusters, we use the KMeans clustering technique on the articles. We’ll also try to label these clusters by selecting the most frequent words that appear in each cluster’s articles.
from cartodata.clustering import create_kmeans_clusters # noqa
n_clusters = 8
def find_clusters(n_clusters, article_embeddings, word_embeddings,
words_matrices, words_scores, matrices):
c_scores_all = []
c_umap_all = []
for i in range(6):
cluster_labels = []
c_lda, c_umap, c_scores, c_knn, _, _, _ = create_kmeans_clusters(n_clusters, # number of clusters to create
# 2D matrix of articles
article_embeddings[i].T,
# the 2D matrix of words
word_embeddings[i].T,
# the articles to words matrix
words_matrices[i],
# word scores
words_scores[i],
# a list of initial cluster labels
cluster_labels,
# space matrix of words
matrices[i][2])
c_scores_all.append(c_scores)
c_umap_all.append(c_umap)
return c_scores_all, c_umap_all
c_scores_all, c_umap_all = find_clusters(n_clusters, article_embeddings, word_embeddings,
words_matrices, words_scores, doc2vec_matrices)
Let’s plot with the clusters.
for i, (embedding_article, embedding_auth, embedding_word, embedding_lab,
color_article, color_auth, color_word, color_lab, c_scores, c_umap) in enumerate(zip(
article_embeddings_names, auth_embeddings_names, word_embeddings_names, lab_embeddings_names,
colors_articles, colors_auth, colors_words, colors_lab, c_scores_all, c_umap_all)):
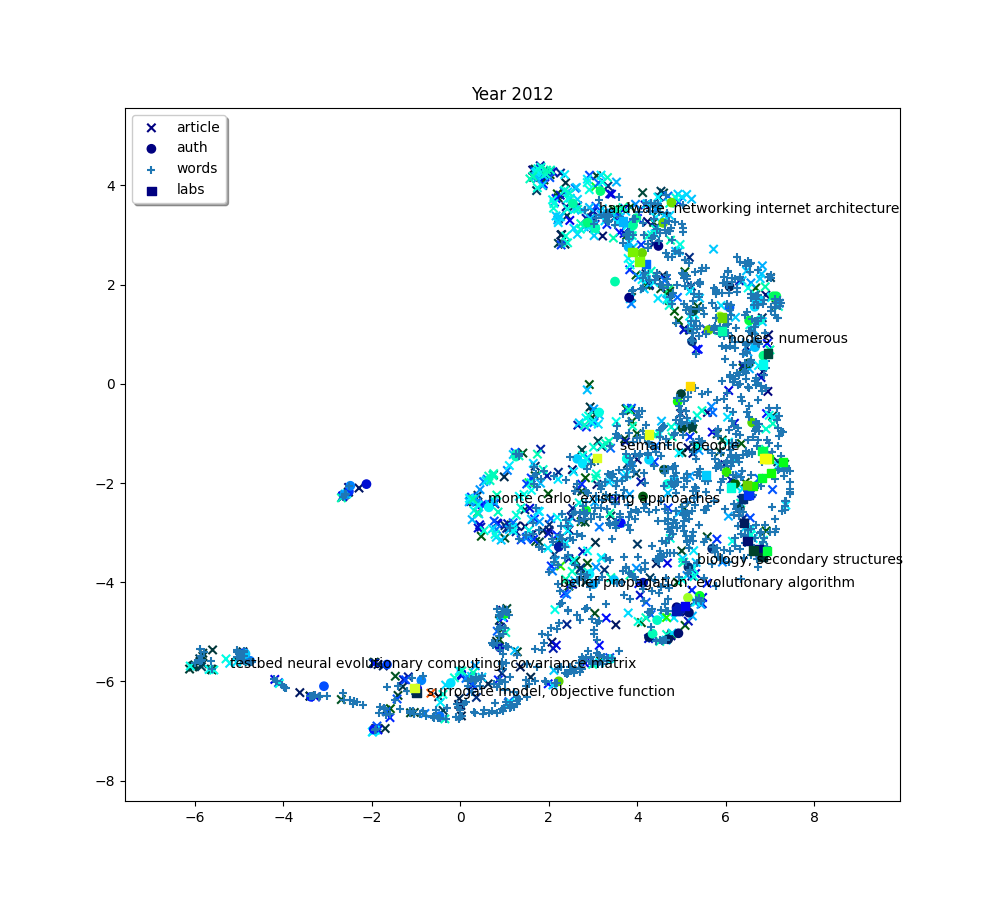
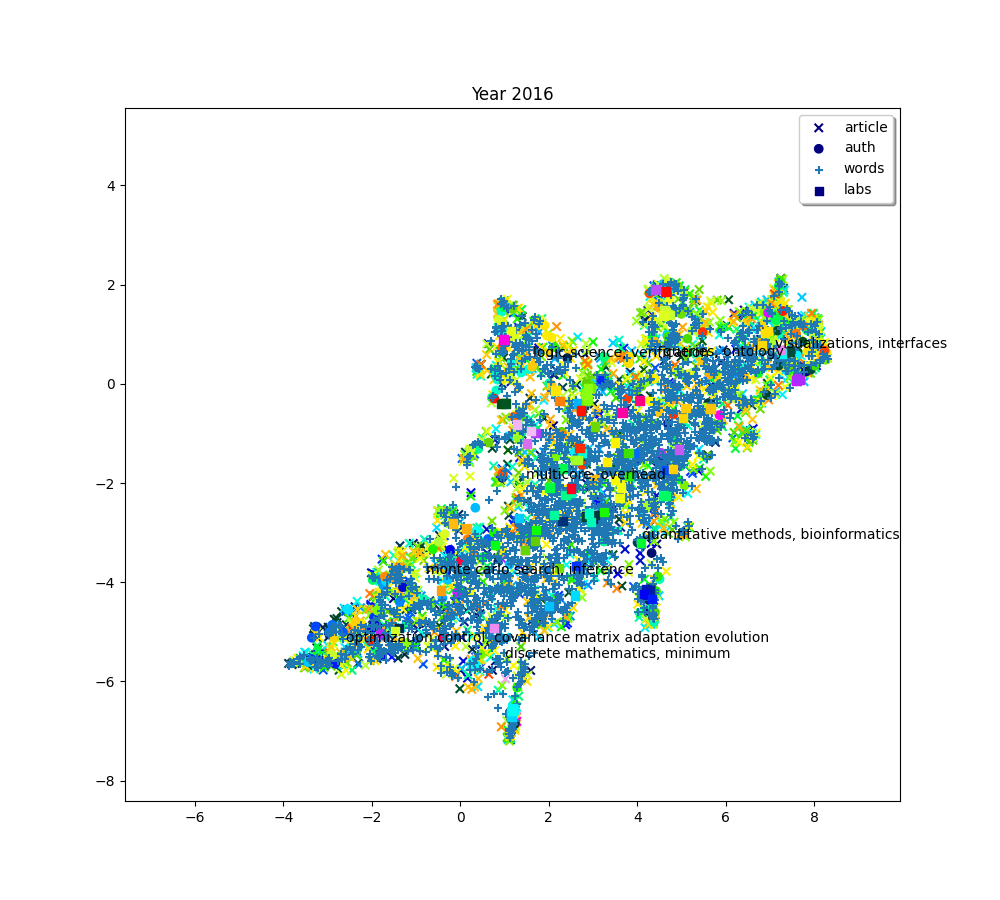
plot(f"Year {2010+ (i*2) + 2}", embedding_article, embedding_auth, embedding_word, embedding_lab,
color_article, color_auth, color_word, color_lab, c_scores, c_umap, n_clusters)
/usr/local/lib/python3.9/site-packages/mplcursors/_pick_info.py:55: UserWarning: No data for colormapping provided via 'c'. Parameters 'vmin', 'vmax' will be ignored
paths = scatter.__wrapped__(*args, **kwargs)
The 8 clusters that we created give us a general idea of what the big clusters of data contain. But we’ll probably want a finer level of detail if we start to zoom in and focus on smaller areas. So we’ll also create a second bigger group of clusters. To do this, simply increase the number of clusters we want.
n_clusters = 32
mc_scores_all, mc_umap_all = find_clusters(
n_clusters, article_embeddings, word_embeddings, words_matrices, words_scores, doc2vec_matrices
)
Warning: Less than 2 words in cluster 5 with (0) words.
Warning: Less than 2 words in cluster 5 with (0) words.
Warning: Less than 2 words in cluster 10 with (0) words.
Warning: Less than 2 words in cluster 10 with (0) words.
Warning: Less than 2 words in cluster 29 with (1) words.
Warning: Less than 2 words in cluster 29 with (1) words.
Warning: Less than 2 words in cluster 8 with (0) words.
Warning: Less than 2 words in cluster 8 with (0) words.
Nearest neighbors¶
One more thing which could be useful to appreciate the quality of our data would be to get each point’s nearest neighbors. If our data processing is done correctly, we expect the related articles, labs, words and authors to be located close to each other.
Finding nearest neighbors is a common task with various algorithms aiming to solve it. The get_neighbors method uses one of these algorithms to find the nearest points of each type. It takes an optional weight parameter to tweak the distance calculation to select points that have a higher score but are maybe a bit farther instead of just selecting the closest neighbors.
Because we want to find the neighbors of each type (articles, authors, words, labs) for all of the entities, we call the get_neighbors method in a loop and store its results in an array.
from cartodata.neighbors import get_neighbors # noqa
def find_neighbors(articles_scores, authors_scores, words_scores,
labs_scores, matrices):
weights = [0, 0.5, 0.5, 0]
all_neighbors = []
all_scores = []
for i in range(6):
scores = [articles_scores[i], authors_scores[i],
words_scores[i], labs_scores[i]]
neighbors = []
matrix = matrices[i]
for idx in range(len(matrix)):
neighbors.append(get_neighbors(matrix[idx],
scores[idx],
matrix,
weights[idx])
)
all_neighbors.append(neighbors)
all_scores.append(scores)
return all_neighbors, all_scores
all_neighbors, all_scores = find_neighbors(scores_article, filtered_auth_scores,
words_scores, filtered_lab_scores, doc2vec_matrices)
Exporting¶
We now have sufficient data to create a meaningfull visualization.
from cartodata.operations import export_to_json # noqa
natures = ['articles',
'authors',
'words',
'labs',
'hl_clusters',
'ml_clusters']
def export(from_year, struct, article_embeddings, authors_embeddings, word_embeddings, lab_embeddings,
c_umap, mc_umap, c_scores, mc_scores, all_neighbors, all_scores):
for i in range(6):
export_file = f"../datas/{struct}_workflow_doc2vec_aligned_{from_year}_{from_year + (2 + 2 * i)}.json"
# add the clusters to list of 2d matrices and scores
umap_matrices = [article_embeddings[i].T,
authors_embeddings[i].T,
word_embeddings[i].T,
lab_embeddings[i].T,
c_umap[i],
mc_umap[i]]
all_scores[i].extend([c_scores[i], mc_scores[i]])
# create a json export file with all the infos
export_to_json(natures,
umap_matrices,
all_scores[i],
export_file,
neighbors_natures=natures[:4],
neighbors=all_neighbors[i])
export(2010, "lisn", article_embeddings, auth_embeddings, word_embeddings, lab_embeddings,
c_umap_all, mc_umap_all, c_scores_all, mc_scores_all, all_neighbors, all_scores)
This creates the files:
lisn_workflow_doc2vec_aligned_2010_2012.json
lisn_workflow_doc2vec_aligned_2010_2014.json
lisn_workflow_doc2vec_aligned_2010_2016.json
lisn_workflow_doc2vec_aligned_2010_2018.json
lisn_workflow_doc2vec_aligned_2010_2020.json
lisn_workflow_doc2vec_aligned_2010_2022.json
each of which contains a list of points ready to be imported into Cartolabe. Have a look at it to check that it contains everything.
import json # noqa
export_file = "../datas/lisn_workflow_doc2vec_aligned_2010_2022.json"
with open(export_file, 'r') as f:
data = json.load(f)
data[1]
{'position': [-0.966591477394104, -4.021040916442871], 'score': 1.0, 'rank': 1, 'nature': 'articles', 'label': 'Bandit-Based Genetic Programming with Application to Reinforcement Learning', 'neighbors': {'articles': [1, 1869, 1478, 834, 387, 1552, 2871, 1895, 2155, 422], 'authors': [2987, 3028, 3043, 3032, 3035, 3053, 3034, 3208, 3157, 3047], 'words': [5394, 3767, 5395, 6515, 3768, 5396, 3652, 4068, 6338, 5651], 'labs': [6944, 7011, 7032, 7066, 6985, 6951, 7030, 6983, 6994, 6996]}}
Total running time of the script: (24 minutes 33.673 seconds)